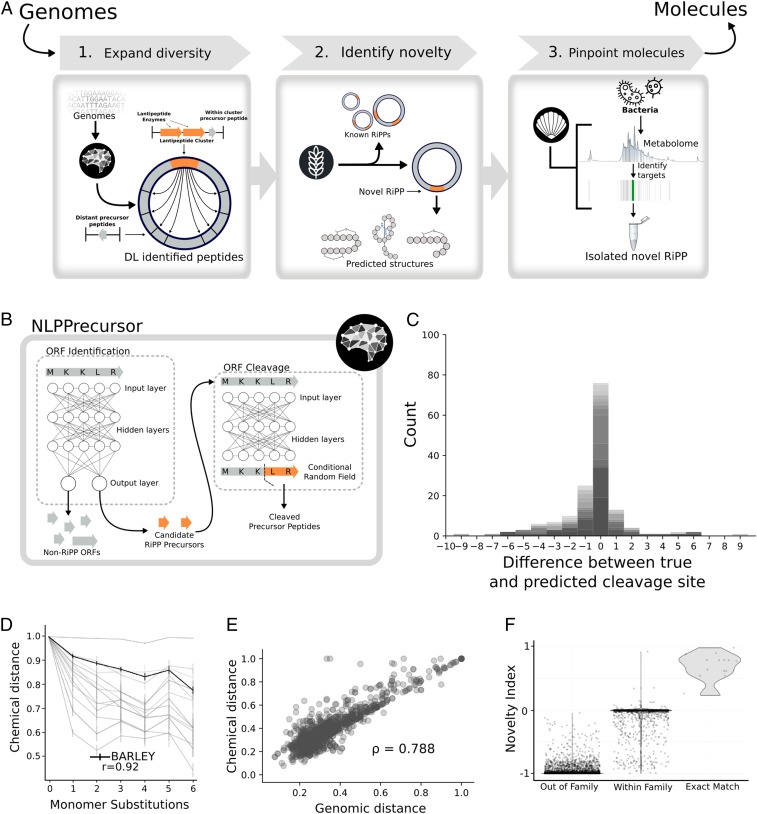
Fig. 2.
Illustration of the DeepRiPP gene to molecule workflow and performance of its genomic modules, NLPPrecursor and BARLEY. (A) The DeepRiPP workflow that guides the discovery strategy from genomes to isolated molecules. DeepRiPP consists of 3 modules. Module 1, NLPPrecursor, implements deep learning techniques inspired by natural language processing to expand the diversity of genomically detected RiPPs by including all potential precursor peptides outside putative biosynthetic gene cluster boundaries. Module 2, BARLEY, identifies novel RiPPs by aligning genomic information to a database of known RiPP chemical structures and scoring the novelty of each candidate RiPP identified by genome mining. Module 3, CLAMS, identifies putative RiPPs in metabolomics data. (B) The architecture of NLPPrecursor, highlighting the 2 components responsible for precursor identification and cleavage, respectively. (C) Histogram depicting the prediction accuracy of NLPPrecursor ORF cleavage, where the x axis is the difference between the predicted and true cleavage site in number of amino acids. Gray shading represent different families. (D) Line chart describing the relationship between increasing chemical divergence in an artificially generated, combinatorial dataset (33) of 600 compounds to chemical distance scores. BARLEY is highlighted in black, while other metrics are shown in light gray. The relationship between the number of monomer substitutions and the chemical similarity assigned by each metric is computed using the Spearman rank correlation coefficient (ρ). (E) Scatterplot representing the relationship between BARLEY chemical distances and genomic distances generated by BARLEY. The comparison was performed on a dataset of 136 known RiPP clusters which encode 161 small molecules (Dataset S3). The Spearman rank correlation coefficient (ρ) is used to quantify the relationship between genomic and chemical BARLEY distances. (F) Validation of BARLEY novelty index. A violin plot is shown with BARLEY predicted novelty index (y axis) and the true relationship type (exact match, family match, or out of family) between encoded RiPP and chemical scaffold (x axis). Using a cutoff of 0.2 on the BARLEY novelty index yields a 99.7% accuracy in classifying exact matches from other comparison types.