Fig. 3.
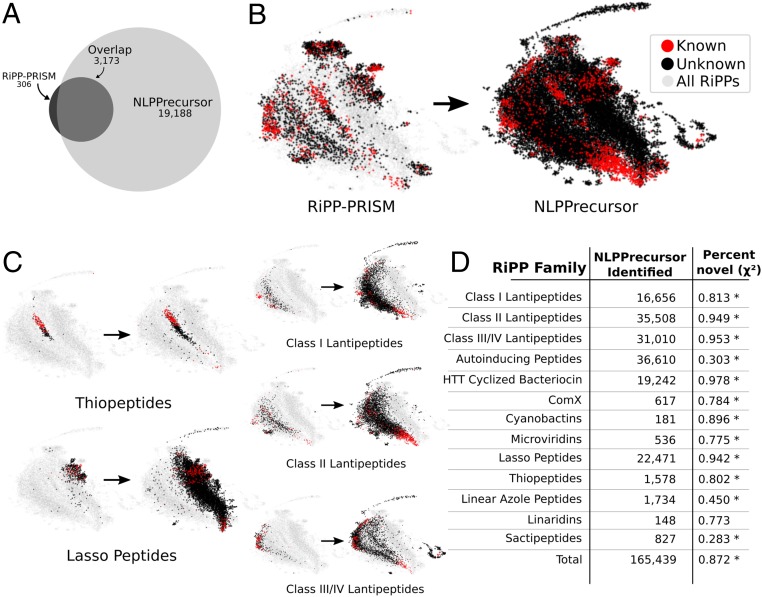
DeepRiPP expands the discovery of novel RiPPs in a reanalysis of genomes analyzed by RiPP-PRISM. (A) Venn diagram depicts the total number of unique RiPP precursor peptides identified via NLPPrecursor as compared to RiPP-PRISM. A total of 65,421 bacterial genomes were analyzed from National Center for Biotechnology Information (downloaded March 2016) through DeepRiPP. (B) Total diversity of genomically encoded RiPPs as identified by RiPP-PRISM and NLPPrecursor distributed according to BARLEY similarity and subsequently plotted using UMAP (36). Each point represents a unique RiPP as determined via BARLEY and is colored according to its novelty as determined by BARLEY. All RiPPs identified by either RiPP-PRISM or NLPPrecursor are shaded in gray within the background to visualize overall localization. (C) Depicting the family-wise increase in diversity using UMAP. (D) Number of RiPPs identified by NLPPrecursor and the percentage of these that are novel. The χ2 test is used to determine whether a significant increase in the percentage novel are observed within the NLPPrecursor set (*P < 10−10).