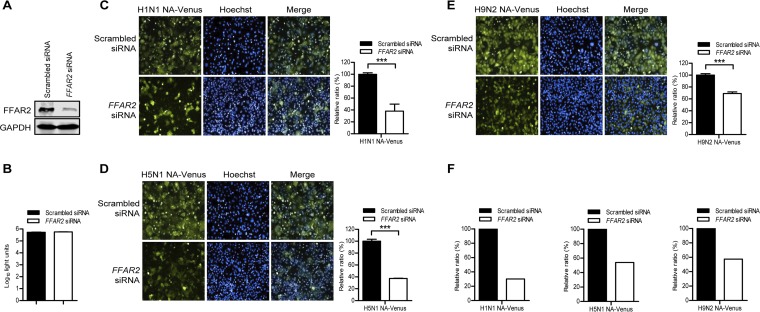
FIG 1.
Identification of FFAR2 as a host factor involved in the replication of IAV by using H1N1 NA-Venus, H5N1 NA-Venus, and H9N2 NA-Venus reporter viruses. (A) A549 cells were transfected with siRNA targeting FFAR2 or with scrambled siRNA for 48 h, and the knockdown of FFAR2 was detected by Western blotting. (B) A549 cells were treated with FFAR2 siRNA or scrambled siRNA and cultured at 37°C for 48 h. Cell viability was determined by using a CellTiter-Glo assay. (C to E) A549 cells were seeded into 96-well plates containing FFAR2 siRNA or scrambled siRNA and cultured at 37°C for 48 h. They were then infected with one of the Venus reporter viruses, H1N1 NA-Venus (MOI = 0.1) (C), H5N1 NA-Venus (MOI = 0.1) (D), or H9N2 NA-Venus (MOI = 0.1) (E), and cultured at 37°C for 24 h. Venus expression was visualized by using the Operetta high-content imaging system. The effect of FFAR2 knockdown by siRNA interference on virus replication was calculated by normalizing the average fluorescence intensity of the FFAR2 siRNA-treated wells with that of the scrambled siRNA-treated cells. The data are presented as means ± standard deviations (SD) for triplicate samples. ***, P < 0.001. (F) A549 cells were infected as described above for panels C to E, and viral NP was detected by immunostaining. The cell nuclei were stained with Hoechst 33342. The blue and red fluorescence images were collected from the same area and used to evaluate the effect of FFAR2 knockdown on the replication of H1N1 NA-Venus, H5N1 NA-Venus, or H9N2 NA-Venus by quantifying the percentage of infected cells as follows: number of cells expressing NP protein (red)/total number of cells (blue).