The efficacy of HIV-1 broadly neutralizing antibody (bnAb) therapies may be compromised by the preexistence of less susceptible variants. Sequence-based methods are needed to predict pretreatment variants’ neutralization sensitivities. HIV-1 strains that exclusively use the CXCR4 receptor rather than the CCR5 receptor are less neutralization susceptible, especially to variable loop 3 (V3 loop) bnAbs in some, but not all, instances. While the inability to utilize the CCR5 receptor maps to a predicted protrusion in the envelope V3 loop, this viral determinant does not directly influence V3 loop bnAb sensitivity. Homology modeling predicts that contact between the envelope V1 loop and the antibody impacts V3 loop bnAb susceptibility in some cases. Among pretreatment envelopes, increased probability of using CXCR4 and greater predicted V1 interference are associated with faster virus rebound and a smaller decrease in the plasma virus level, respectively, after V3 loop bnAb treatment. Receptor usage information and homology models may be useful for predicting V3 loop bnAb therapy efficacy.
KEYWORDS: cell envelope, human immunodeficiency virus, neutralizing antibodies, receptors, surface structures
ABSTRACT
In clinical trials, HIV-1 broadly neutralizing antibodies (bnAbs) effectively lower plasma viremia and delay virus reemergence. The presence of less neutralization-susceptible strains prior to treatment decreases the efficacy of these antibody-based treatments, but neutralization sensitivity often cannot be predicted by sequence analysis alone. We found that phenotypically confirmed CXCR4-utilizing strains are less neutralization sensitive, especially to variable loop 3 (V3 loop)-directed bnAbs, than exclusively CCR5-utilizing strains in some, but not all, cases. Homology modeling suggested that the primary V3 loop bnAb epitope is equally accessible among CCR5- and CXCR4-using strains, although variants that exclusively use CXCR4 have V3 loop protrusions that interfere with CCR5 receptor interactions. Homology modeling also showed that among some, but not all, envelopes with decreased neutralization sensitivity, V1 loop orientation interfered with V3 loop-directed bnAb binding. Thus, there are likely different structural reasons for the coreceptor usage restriction and the different bnAb susceptibilities. Importantly, we show that individuals harboring envelopes with higher likelihood of using CXCR4 or greater predicted V1 loop interference have faster virus rebound and a lower maximum decrease in plasma viremia, respectively, after treatment with a V3 loop bnAb. Knowledge of receptor usage and homology models may be useful in developing future algorithms that predict treatment efficacy with V3 loop bnAbs.
IMPORTANCE The efficacy of HIV-1 broadly neutralizing antibody (bnAb) therapies may be compromised by the preexistence of less susceptible variants. Sequence-based methods are needed to predict pretreatment variants’ neutralization sensitivities. HIV-1 strains that exclusively use the CXCR4 receptor rather than the CCR5 receptor are less neutralization susceptible, especially to variable loop 3 (V3 loop) bnAbs in some, but not all, instances. While the inability to utilize the CCR5 receptor maps to a predicted protrusion in the envelope V3 loop, this viral determinant does not directly influence V3 loop bnAb sensitivity. Homology modeling predicts that contact between the envelope V1 loop and the antibody impacts V3 loop bnAb susceptibility in some cases. Among pretreatment envelopes, increased probability of using CXCR4 and greater predicted V1 interference are associated with faster virus rebound and a smaller decrease in the plasma virus level, respectively, after V3 loop bnAb treatment. Receptor usage information and homology models may be useful for predicting V3 loop bnAb therapy efficacy.
INTRODUCTION
Multiple broadly neutralizing antibodies (bnAbs) are being examined as novel therapeutics against human immunodeficiency virus type 1 (HIV-1) infection (1–6). In contrast to the current highly effective antiretroviral drugs (ARVs), antibody-based therapies require less frequent dosing, can be effective against drug-resistant variants, and may potentiate host humoral responses (7). Prior to initiating ARVs, HIV-1-infected patients are routinely evaluated for the presence of drug-resistant strains, primarily using sequence-based methods (8). BnAb sensitivity has also been associated with specific HIV-1 envelope (Env) sequences. For V3 loop-directed bnAbs, modifications to the epitope, the asparagine (N)-linked glycan at Env position 332 (N332), impacts susceptibility (9–13). A recent study demonstrated that V1-V2 loop characteristics, such as net charge and length, are also associated with sensitivity (13). To date, however, clinical trials have used phenotype-based methods to identify pretreatment variants with reduced bnAb susceptibility, although the assays are cumbersome and lack sensitivity (2). Greater insight into Env characteristics that are associated with bnAb susceptibility may be helpful in developing better tests.
In addition to being antibody targets, the Env V1-V2 and V3 loops also influence binding to either the CCR5 or CXCR4 coreceptor, and this attachment is necessary for host cell entry (14–16). We and others have shown that variants that use the CXCR4 receptor (termed X4 or R5X4) are less sensitive to V1-V2 and V3 loop bnAbs than those that use only CCR5 (classified as R5) (17–19). This association potentially exists because the glycan domain that contains V1-V2 and V3 loop bnAb epitopes shields the CCR5 coreceptor binding site from antibodies (19, 20). As opposed to sequence features, structure-based models have been used to predict CCR5 and CXCR4 usage with greater accuracy (21). Similar methods could potentially be used to speculate about bnAb sensitivity and its association with coreceptor usage. Furthermore, envelope-antibody homology models may also provide novel insights, which could be helpful in predicting bnAb susceptibility in the future.
(This article was submitted to an online preprint archive [22].)
RESULTS
X4 variants are less susceptible to neutralization by plasma- and glycan-directed bnAbs.
We have shown that HIV-1 subtype C (HIV-1C) X4 variants are more neutralization resistant to autologous contemporaneous plasma than cocirculating R5X4 and R5 strains (17). Some earlier studies, however, implied that HIV-1B CXCR4-using Envs are more neutralization sensitive than CCR5-using Envs (23–27). We compared the neutralization sensitivities of HIV-1B variants to examine this question in further detail. We isolated a total of 929 individual Envs (median, 16 Envs per subject; range, 1 to 239) using single-genome amplification (SGA) from 33 antiretroviral-naive patient samples previously classified as dual mixed (DM), i.e., having either X4 or R5X4 strains (28, 29). A Web-based prediction tool, either WebPSSM or Geno2Pheno, was used to predict the coreceptor usage of the isolated Envs based on the V3 loop sequence (30, 31). Some of the SGA Envs from 22 individuals were genotypically predicted to use CXCR4, and the predicted CXCR4-utilizing Envs were likely not isolated from the other 11 previously characterized DM samples because of relatively limited SGA sampling compared to the bulk PCR analysis. Some of the isolated Envs from 17 of the 22 samples were examined for coreceptor usage by incorporating them into an HIV-1 backbone with the isogenic envelope deleted (Table 1). In three (4102, 1239, and 1924) samples, we were able to isolate cocirculating R5 and X4 strains, while two others (3248 and 3576) yielded X4 Envs only. The isolated SGA Envs examined from the remaining 12 samples were either all R5, even though some of them were predicted by sequencing to use the CXCR4 receptor, or a mixture of R5X4 and R5 (Table 1).
TABLE 1.
Subject envelope characteristics
| Subject | No. of SGA Envs isolated | No. of SGA Envs with predicted genotype |
No. of Envs phenotyped | No. of recombinant viruses with confirmed phenotype |
|||
|---|---|---|---|---|---|---|---|
| R5 | CXCR4 | R5 | X4 | R5/X4 | |||
| 4102 | 61 | 52 | 9 | 19 | 13 | 5 | 1 |
| 1239 | 239 | 234 | 5 | 12 | 9 | 1 | 2 |
| 3248 | 16 | 0 | 16 | 9 | 0 | 9 | 0 |
| 1924 | 27 | 24 | 3 | 2 | 1 | 1 | 0 |
| 3576 | 30 | 0 | 30 | 6 | 0 | 6 | 0 |
| 3131 | 17 | 6 | 11 | 7 | 7 | 0 | 0 |
| 1069 | 10 | 0 | 10 | 9 | 9 | 0 | 0 |
| 2327 | 6 | 0 | 6 | 4 | 1 | 0 | 3 |
| 0229 | 40 | 5 | 35 | 7 | 3 | 0 | 4 |
| 1045 | 15 | 5 | 10 | 5 | 5 | 0 | 0 |
| SC | 11 | 7 | 4 | 6 | 5 | 0 | 1 |
| 1389 | 14 | 0 | 14 | 7 | 1 | 0 | 5 |
| 1486 | 15 | 7 | 8 | 5 | 5 | 0 | 0 |
| 3026 | 110 | 109 | 1 | 4 | 4 | 0 | 0 |
| 1233 | 98 | 72 | 26 | 10 | 7 | 0 | 0 |
| 9265 | 14 | 0 | 14 | 14 | 0 | 0 | 14 |
| 1874 | 11 | 2 | 9 | 5 | 5 | 0 | 0 |
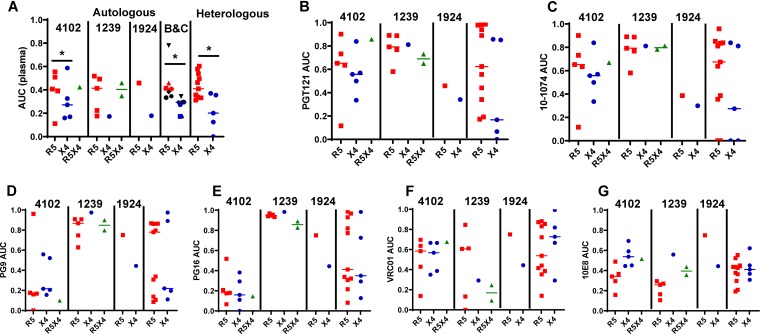
Neutralization sensitivity was estimated using the area under the curve (AUC) rather than the concentration required to achieve 50% inhibition (IC50), because the two estimates are highly correlated and the AUC can be used when 50% inhibition is not observed at the highest tested concentration (32, 33). Neutralization sensitivities to autologous contemporaneous plasma were compared for up to a maximum of five unique cocirculating 4102, 1239, and 1924 R5, R5X4, and X4 strains. A statistically significant difference was observed only between 4102 X4 and R5 variants (P = 0.01) and not for R5X4, 1239, or 1924 strains because of limited numbers of isolates (Fig. 1A). The small number of individuals with cocirculating R5 and X4 variants limited the ability to draw firm conclusions. To overcome this issue, we combined the data from our previous examination of 4 individuals infected with cocirculating HIV-1C R5 and X4 strains (17). In an aggregate HIV-1B and HIV-1C assessment, X4 strains were significantly more resistant to contemporaneous autologous plasma than cocirculating R5 strains (P = 0.02; n = 7 individuals) (Fig. 1A). The plasma neutralization sensitivities of a group of primary X4 and R5 Envs composed of one randomly selected HIV-1B X4 (n = 5) and R5 (n = 11) variant from different individuals were also compared (see Materials and Methods). Susceptibility to a standard comparator generated by pooling plasma from 10 HIV-1B-infected individuals, different from the 17 subjects mentioned above, was assessed. The HIV-1B X4 group was around 2-fold less sensitive to heterologous plasma than the R5 group (P = 0.03) (Fig. 1A).
FIG 1.
X4 strains are less susceptible than R5 strains to plasma- and V3-directed bnAbs. Shown are neutralization AUCs (y axes) among primary R5 (red), X4 (blue), and R5X4 (green) Envs against autologous contemporaneous and heterologous plasma (A), PGT121 (B), 10-1074 (C), PG9 (D), PG16 (E), VRC01 (F), and 10E8 (G). Each point denotes a unique Env, and the values represent means from duplicate independent experiments. In all the dot plots, the lines indicate medians. The subject IDs identify cocirculating strains. In panel A, the dot plot labeled B&C shows the median AUCs of cocirculating variants from 3 HIV-1B (red and blue) and 4 HIV-1C (black) strains against autologous plasma. Different symbols represent different subjects. In each panel, the rightmost dot plot is a comparison between unrelated variants. Comparisons were done using nonparametric Wilcoxon rank sum tests. *, P < 0.05.
A previous study examined differences of neutralization sensitivities to first-generation, but not second-generation, bnAbs among cocirculating HIV-1B variants (26). First-generation bnAbs do not target the V3 loop. We were interested in examining susceptibility to second-generation bnAbs because our previous investigation suggested that in some cases emergence of CXCR4-using strains might be due V3-directed antibodies (17). In two cases (4102 and 1924), but not in another case (1239), X4 variants were more sensitive to N332 glycan-directed bnAbs (PGT121 and 10-1074) than cocirculating R5 variants, although the differences were not statistically significant (Fig. 1B and C). Thus, similar to HIV-1C, in some but not all cases, X4 strains were more resistant to V3 loop-directed bnAbs than cocirculating R5 strains (17). In contrast to cocirculating variants, the X4 variants were around 4- to 8-fold less sensitive to the V3-directed antibodies PGT121 and 10-1074 than R5 variants from different subjects, although the differences only showed a statistical trend, likely due to the small sample size (P = 0.09 for both) (Fig. 1B and C).
Both cocirculating and X4 and R5 Envs from different subjects showed similar susceptibilities to V1-V2 (PG9 and PG16)-directed, CD4 binding site (bs) (VRC01)-directed, and membrane-proximal external-region (MPER)-directed (10E8) bnAbs (Fig. 1D to G). These observations were confirmed by comparing the neutralization sensitivities of phenotypically confirmed CXCR4-using variants (either X4 or R5X4) to those of R5 strains in the Los Alamos CATNAP database (34). A larger proportion of the CXCR4-using variants than of R5 variants had IC50s above the highest tested bnAb concentration for the V3-directed glycan bnAb, PGT121 (P < 0.0001) and the V1-V2 glycan-dependent bnAbs, PG9 (P = 0.02) and PG16 (P = 0.03), but not for the CD4 bs bnAb, VRC01 (P = 0.95). The tested but undetectable IC50s were given a value of 100 μg/ml for the subsequent statistical comparisons. The CXCR4-utilizing variants (median, 100 μg/ml; range, 0.009 to 100 μg/ml; n = 20) had significantly higher IC50s to PGT121 than the R5 strains (median, 0.10 μg/ml; range, 0.001 to 100 μg/ml; n = 134; P = 0.0002). The R5 variants (median PG9 IC50, 0.26 μg/ml; range, 0.002 to 100 μg/ml; n = 135; median PG16 IC50 0.25 μg/ml; range, 0.009 to 100 μg/ml; n = 131) also had significantly lower IC50s to PG9 and PG16 than the CXCR4-using strains (median, PG9 IC50, 1.96 μg/ml; range, 0.006 to 100 μg/ml; n = 21; P = 0.03; median PG16 IC50, 34.35 μg/ml; range, 0.001 to 100 μg/ml; n = 21; P = 0.04). On the other hand, CXCR4-using variants (median, 0.55 μg/ml; range, 0.04 to 100 μg/ml; n = 22) and R5 (median, 0.50 μg/ml; range, 0.01 to 100 μg/ml; n = 149) had similar neutralization susceptibilities to VRC01 (P = 0.26). A small number of R5X4 variants, both in the CATNAP database and in our primary Env analysis, precluded their examination as an independent group. Similar to previous studies, in general, X4 Envs are less neutralization susceptible than R5 Envs, especially to V3 loop-directed bnAbs, but this is not always true among cocirculating X4 and R5 strains (18, 19, 34).
Plasmas that have CXCR4-using strains do not have greater neutralization capacity than those with only CCR5-using strains.
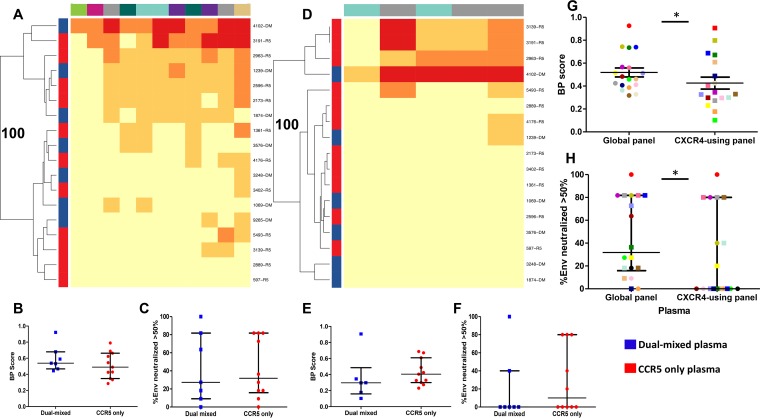
HIV-1-infected individuals generate new antibodies to neutralize the viruses, which evolve over time during chronic infection (35, 36). We hypothesized that individuals with X4 or R5X4 strains may have greater plasma neutralization capacity than those with R5-only variants because more potent antibodies would evolve over time in response to the presence of relatively neutralization-insensitive CXCR4-using Envs. We tested this prediction by comparing neutralization breadths and potencies among plasmas previously classified as R5 only (n = 11) or as DM (n = 7). This classification was primarily based on previously described bulk Env genotype and phenotype characterization (29), which is standard in the field, even though it is not perfect. In some cases, isolated SGA Envs were used to confirm the classification. Although the duration of infection was unknown, as expected, the individuals with R5-only populations had significantly higher absolute CD4 counts (P = 0.04) than those with DM populations, but no significant difference in plasma virus levels (P = 0.19) (37, 38). The ability of a plasma sample to neutralize a global reference panel of 11 R5 Envs of various subtypes was used to estimate a neutralization breadth and potency (BP) score. This method has been previously validated as a measure of neutralization capacity (33). Heat maps depicting the neutralization responses against the 11 Envs revealed that plasmas from individuals from the two groups were not qualitatively different (Fig. 2A). The DM and R5 plasmas had similar BP scores (P = 0.37) (Fig. 2B) and neutralized similar percentages of the 11-Env reference panel at greater than 50% (P = 0.88) (Fig. 2C). Although the sample size is relatively small, there was 80% power, at a type 1 error level of 0.05, to detect around 1.5-fold or greater differences in the median BP scores based on the observed distributions.
FIG 2.
Samples containing CXCR4-using viruses and those with CCR5-only viruses have similar neutralization potencies and breadths, but plasmas have decreased ability to neutralize CXCR4-using compared to R5 strains. (A and D) Heat maps showing plasma neutralization against the R5 global reference Env panel (A) and a CXCR4-using Env collection (D). Each square in the heat map represents the average percent neutralization for the Env-plasma combination tested: yellow, <50%; light orange, 50 to 70%; dark orange, 70 to 90%; red, >90%. On the left, blue and red denote DM and R5-only plasmas, respectively, and individual plasma IDs are listed on the right. Env subtypes are indicated by color above the heat maps: A, khaki; B, gray; C, teal; G, green; AC, pink; CRF01_AE, dark green; and CRF07_BC, purple. The branches show the hierarchical clustering, with bootstrap probability for 100 iterations. (B and E) BP scores for DM (blue) and CCR5-only (red) plasmas against the global Env panel (B) and a CXCR-using Env collection (E). (C and F) Breadths (percentage of Envs neutralized at greater than 50% at the highest tested plasma dilution) observed for the DM (blue) and CCR5-only (red) plasmas against the global Env panel (C) and a CXCR4-using Env collection (F). Comparisons were done using the Wilcoxon rank sum test. (G and H) Plasma BP scores (G) and breadths (H) against the global Env panel compared to the CXCR4-using Env panel. Each unique plasma sample is denoted by a different color/symbol. Comparisons were done using a matched-pairs Wilcoxon rank sum test. In all the box plots, the values are means from a minimum of 2 independent assays; the lines denote medians and interquartile ranges. *, P < 0.05.
The global reference panel contains only R5 Envs, and therefore, neutralization capacity was also examined against another Env collection consisting of a relatively small number (n = 5) of CXCR4-using variants (14, 39, 40). DM and R5-only plasmas had similar neutralization fingerprints against this CXCR4-using Env collection (Fig. 2D), and there was no significant difference in BP scores (P = 0.22) (Fig. 2E) or breadth (P = 0.83) (Fig. 2F). The DM and R5-only plasmas (n = 18), however, had around 1.5-fold lower BP scores against the CXCR4-using plasmas than against the global reference Env panel (P = 0.01) (Fig. 2G). Furthermore, a significantly lower percentage of the CXCR4-using panel than of the global reference Envs were neutralized by greater than 50% (P = 0.02) (Fig. 2H). This further confirms that CXCR4-using Envs have decreased susceptibility to plasma neutralization compared to R5 Envs. The presence of these less neutralization-susceptible CXCR4-using strains, however, does not necessarily lead to the emergence of different, more potent antibodies.
V3 loop protrusions impact CCR5 receptor interactions, but not access to the V3 loop bnAb epitope.
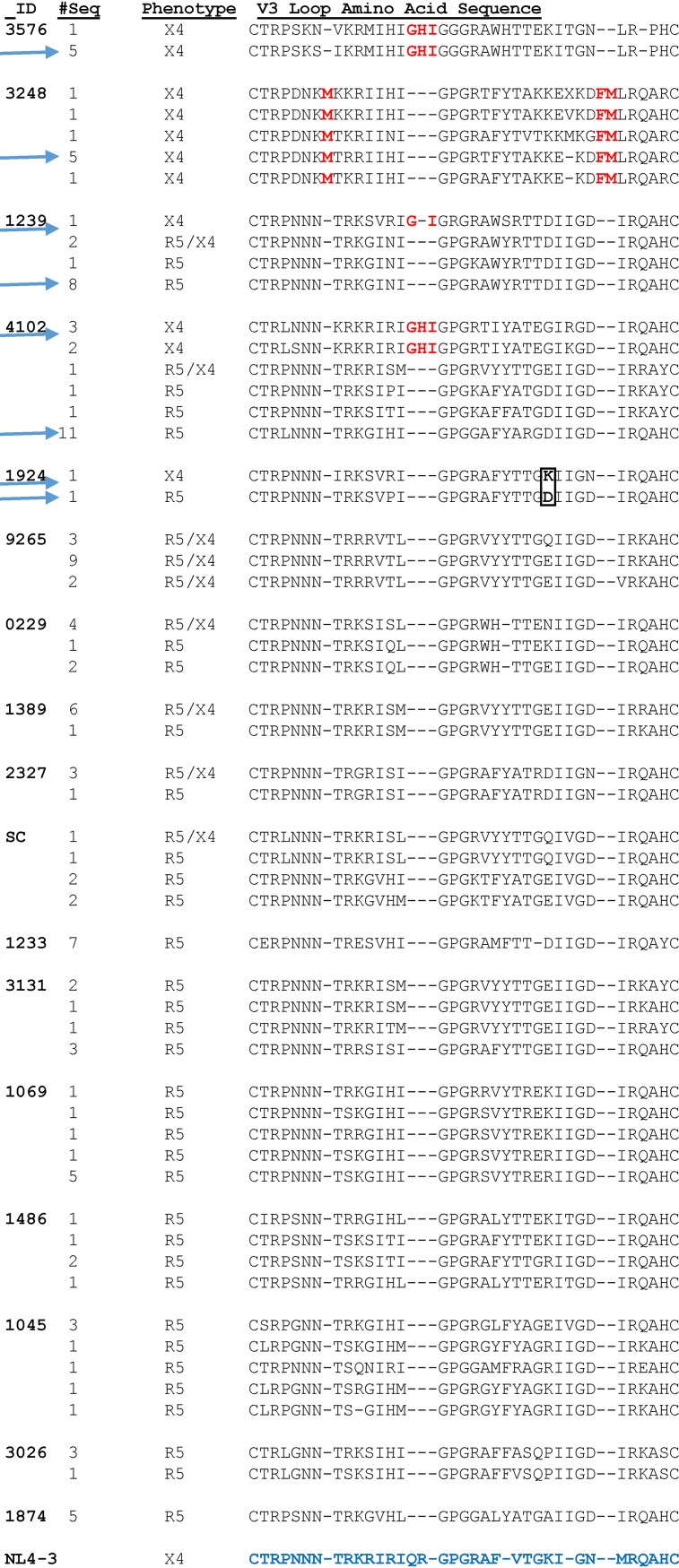
All the R5 and X4 variants whose neutralization susceptibilities to PGT121 and 10-1074 were compared (Fig. 1B and C) had a predicted glycan at Env position 332. A sequence alignment of 22 X4, 31 R5X4, and 77 R5 primary Envs with phenotypically confirmed receptor usage revealed that all the X4 variants except 1924 contained a 2- or 3-amino-acid V3 loop insertion either directly before the glycine (G)-proline (P)-G crown or toward the base of the V3 loop (Fig. 3). These specific V3 modifications were not found in any of the phenotypically confirmed R5 Envs. In contrast, the X4 variant in subject 1924 contained a positively charged amino acid substitution at Env position 321 (V3 loop position 25), which has previously been associated with CXCR4 receptor usage (14, 30).
FIG 3.
A majority of primary X4 Envs contain V3 loop insertions. Shown are alignments of the predicted amino acid sequences of phenotypically confirmed X4, R5, and R5/X4 Env V3 loops from 17 subjects. The columns on the left indicate the subject ID, the number of Envs with the same predicted V3 loop sequence, and the confirmed coreceptor phenotype. The red letters denote insertions. The K and D amino acid difference at V3 loop position 25 in the 1924 X4 and R5 Envs is boxed. The HXB2 (an X4 strain) V3 loop amino acid sequence is at the bottom in blue. The arrows indicate the sequences used for the V3 homology models.
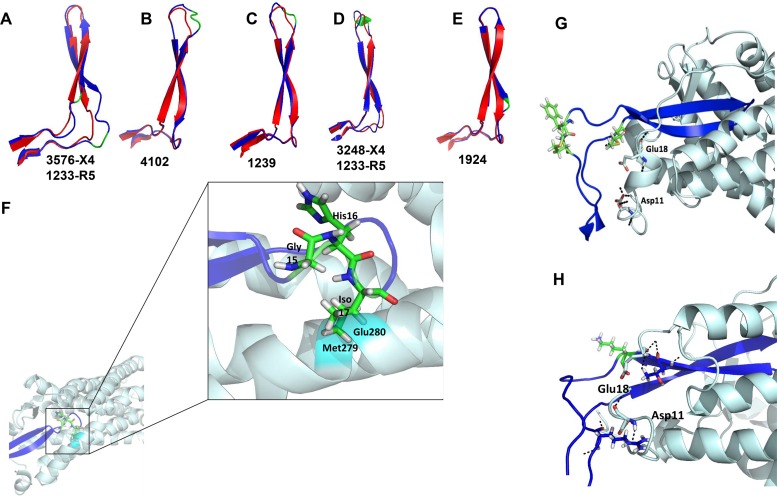
We hypothesized that these observed V3 loop sequence motifs associated with X4 strains restricted coreceptor usage and possibly access to the primary V3 loop bnAb epitope. Structural-homology models were used to assess this premise. The predicted V3 loop structure of X4 Envs (1239, 1924, 4102, 3248, and 3576) was compared to that of either a cocirculating R5 variant (1239, 1924, and 4102) or a heterologous R5 strain (1233). Superimposed Env structures revealed a secondary protrusion in all the phenotypically confirmed X4 V3 loops compared to the R5 V3 loop Envs (Fig. 4A to E). As expected, this protuberance coincided with the location of the insertion at either the tip or the base of the V3 loop. Interestingly, the 1924 X4 V3 loop also contained a protrusion in the V3 loop in the absence of an insertion, which directly corresponded to the observed aspartic acid (D)-to-lysine (K) substitution (Fig. 4E).
FIG 4.
X4 V3 loops contain a protrusion that impairs CCR5 binding. (A to E) Superimposed predicted X4 (blue) and R5 (red) V3 loop structures. The locations of the observed insertions and amino acid substitutions are depicted in green. The ID(s) below each predicted structure indicates the identity of the subject for the X4 V3 loop and either a cocirculating or heterologous R5 V3 loop. (F) Interaction of a predicted 4102-X4 V3 loop structure (purple) on the predicted 4102-R5 V3 loop-CCR5 (light blue) model. Predicted steric clashes at positions 279 and 280 are highlighted in cyan. The stick configuration at the tip of the V3 loop shows the 3-amino-acid insertion (green) observed in the 4102 X4 Env. (G and H) Interaction of 3248 (G) and 1924 (H) X4 V3 loops (purple) with the CCR5 receptor (light blue). The stick configurations show the amino acid insertions (green) observed in the X4 Envs and amino acids Glu18 and Asp11 of the CCR5 receptor. The black dashes represent hydrogen bonds that were absent in the model compared to the 4102-R5 V3 loop and CCR5 structures.
Until recently, there was no solved CCR5 structure in conjunction with the HIV-1 Env (20). We initially used a previous structural model of CCR5 created on a CXCR4 template to examine the impact of the V3 loop protrusion (41). Receptor-ligand interactions for CCR5 and a 4102 R5 V3 loop Env were predicted using ClusPro (42). In these simulations, R5 V3 loops interacted with CCR5 in an expected manner (data not shown) (41). In contrast, X4 interactions with CCR5 could not be examined because all docking simulations put the X4 V3 loop in orientations that did not interact with the CCR5 receptor. Therefore, the X4 V3 loop homology model was superimposed onto the predicted CCR5-4102 R5 V3 loop model complex (Fig. 4F to H). The R5-utilizing V3 loop was then removed for visual clarity. The 4102 X4 Env V3 loop crown insertions clashed with methionine 279 (M279) and glutamic acid 280 (E280) in CCR5 extracellular loop 2 (ECL2) (Fig. 4F). On the other hand, subject 3248 X4 Env with the insertion at the base of the V3 loop eliminated two hydrogen bonds known to be important for CCR5 binding, namely, V3 arginine 3 (R3) to CCR5 aspartic acid 11 (D11) and V3 R23 to CCR5 E18 (Fig. 4G) (41). These two important hydrogen bonds were also not observed for the 1924 X4 V3 loop with the predicted protrusion (Fig. 4H). Similar steric clashes and absence of important hydrogen bonds were also observed using the recently solved HIV-1 Env-CCR5 structure (data not shown). This further confirms the previous conclusion that the V3 loop binding pockets on the CCR5 and CXCR4 receptors are very similar (20).
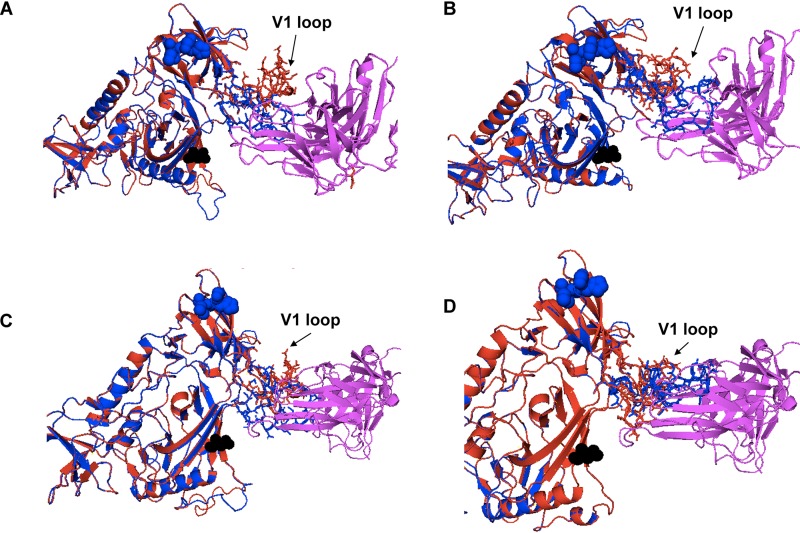
Next, homology models were used to examine how the X4 Env V3 loop insertion-related protrusions impacted V3 loop bnAb access. First, Env structures were predicted using SWISS-MODEL with Env BG505-SOSIP-gp140 as the template (43). Next, within PyMOL (Schrödinger LLC; version 2.2.2), the predicted models were superimposed on the crystal structure of a BG505-SOSIP in complex with either 3H+109L (Protein Data Bank identifier [PDB ID] 5CEZ) or 10-1074 (PDB ID 5T3Z) (44, 45). The PGT121 precursor 3H+109L structure was chosen in the homology modeling because the PGT121 structure has not been solved in conjunction with the Env gp120 subunit. In addition, the epitope binding regions for 3H+109L and PGT121 (PDB ID 4FQ1) have relatively small root mean square deviations (RMSD = 1.37 Å) and similar angles of approach to the Env. Homology models revealed that the targeted V3 loop epitope was equally accessible to the PGT121 precursor and 10-1074 in the relatively resistant insertion containing X4 (4102-3_6 and 4102-3_5) and the relatively sensitive insertion-deficient R5 Envs (4102-61 and 4102-2_17) (Fig. 5A to D). Thus, the homology modeling implied that V3 loop protrusions associated with X4 strains restrict CCR5 receptor, but not V3 loop, bnAb interactions.
FIG 5.
The orientation of the V1 loop influences neutralization sensitivity to anti-V3 loop antibodies. Shown are homology models of 3H+109L, a precursor to PGT121 (magenta) (A and B), and 10-1074 bnAb (magenta) (C and D) interaction with relatively sensitive R5 (4102_61 [A and C] and 4102-2_17 [B and D]) (red) and less susceptible X4 (4102-3_6 [A and C] and 4102-3_5 [B and D]) (blue) Envs. The black spheres in each structure represent the predicted N332 sites. The blue spheres show the V3 loop insertions in the X4 strains compared to the R5 strains. The V1 loop is indicated with arrows.
Contact between the V1 loop and the bnAb impacts susceptibility.
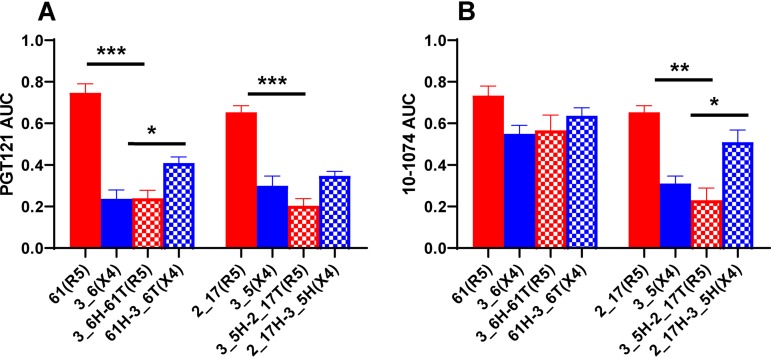
The homology models also revealed that the V1 loop of the highly sensitive Envs, pointed away from the PGT121 precursor and the 10-1074 bnAb (Fig. 5). On the other hand, the V1 loops of the relatively resistant Envs clashed with the antibodies. We used chimeric Envs to confirm the predicted impact of V1-V2 loops on susceptibility to V3 loop-directed bnAbs. V1-V2 loops were swapped from 4102 R5 viruses highly sensitive to PGT121 (4102-61 and 4102-2_17) and X4 variants relatively resistant to the bnAb (4102-3_6 and 4102-3_5). The chimeras contained exchanged domains from the start of the Env gene to the end of V1-V2 (labeled “head” [H]) and from the V1-V2 terminus to the Env end (termed “tail” [T]). Highly susceptible R5 Env PGT121 and 10-1074 sensitivities significantly decreased after the introduction of the X4 head domains in three of four cases (Fig. 6). In contrast, bnAb susceptibility of the relatively resistant Env V3 loop significantly increased after the introduction of the heads from the highly sensitive Envs in two of the four cases. These swaps yielded Envs that were not as highly susceptible or as relatively resistant as the original nonchimeric strains. Furthermore, the exchanges did not switch receptor usage. Chimeras between cocirculating R5 and X4 Envs from other individuals (1239 and 1924) were not produced because of relatively small differences in susceptibility to PGT121 and 10-1074 (Fig. 1B and C). It should be noted that, compared to the original Envs, the chimeric Envs contained exchanged V1-V2 loops, along with some amino acid differences present prior to the start of the variable loops. Thus, it is possible that changes prior to the V1-V2 loop also impact neutralization sensitivity.
FIG 6.
Env V1-V2 loops impact susceptibility to V3 loop-directed antibodies. The graphs show neutralization AUCs for PGT121 (A) and 10-1074 (B) for the original R5 (red bars), original X4 (blue bars), Env with an X4 V1-V2 loop in an R5 background (red crosshatched bars), and Env with an R5 V1-V2 loop in an X4 background (blue crosshatched bars). Below the x axis, the Env clone followed by the observed coreceptor usage in parentheses are shown. The bars show means, and the error bars show standard errors from three independent replicates. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; t test with Welch’s correction.
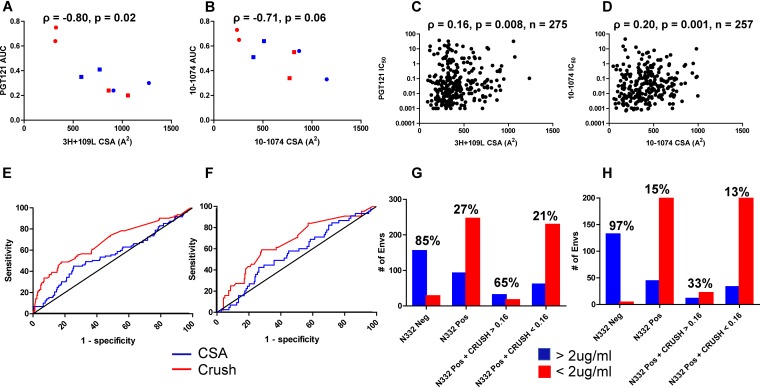
The influence of Env V1 loop orientation was further examined by estimating the contact surface area (CSA) between a predicted Env V1 loop structure and an antibody. In this context, a larger CSA implied greater proximity of the V1 loop to the antibody, and vice versa. Within PyMOL, the CSA was estimated as the sum of the solvent-accessible area for the antibody and V1 loop structure individually minus the solvent-accessible area for the V1 loop in complex with the antibody (46). As expected from the predicted structures (Fig. 5), the relatively resistant X4 (4102-3_6 and 4102-3_5) Envs had greater CSAs than the highly sensitive R5 (4102-61 and 4102-2_17) Envs (Fig. 7A and B). Among the original and chimeric Envs, the estimated CSA increased as the neutralization PGT121 and 10-1074 AUCs decreased (Fig. 7A and B). To further confirm this association, 3H+109L and 10-1074 CSAs were estimated for all the Envs in the CATNAP database with a predicted N332 site and detectable IC50. As the CSA increased, sensitivity to PGT121 and 10-1074 decreased, and the association was statistically significant (Fig. 7C and D). There were a large number of outliers, suggesting that the variable loop has an impact on V3 loop bnAb susceptibility for some, but not all, Envs. In aggregate, the chimeric Env and CATNAP database analyses suggest that the V1 loop impacts susceptibility to V3 loop bnAbs.
FIG 7.
Sequence-dependent coreceptor utilization and contact between the V1 loop and antibody predict neutralization sensitivity. (A and B) Correlation between estimated V1 loop and 3H+109L (A) and 10-1074 (B) CSA (x axes) and PGT121 (A) and 10-1074 (B) neutralization AUCs (y axes) for subject 4102 original R5 (red circles) and X4 (blue circles) Envs and chimeric R5 (red squares) and X4 (blue squares) Envs. (C and D) Correlation between V1 loop and 3H+109L (C) and 10-1074 (D) CSA (x axes) and IC50 (y axes) among all CATNAP Envs with detectable neutralization. The graphs in panels A to D show Spearman rank correlations. (E and F) ROCs showing CRUSH (red) and CSA (blue) in predicting Envs with PGT121 (E) and 10-1074 (F) IC50s of >2 μg/ml versus <2 μg/ml. The black lines are the lines of identity. (G and H) Numbers of Envs (y axes) with the defined characteristics (x axes) that had IC50s of >2 μg/ml (blue) or <2 μg/ml (red) for PGT121 (G) and 10-1074 (H). The percentages of Envs with documented IC50s above 2 μg/ml relative to the total numbers with the characteristics indicated on the x axes are shown above the bars.
Sequence-derived coreceptor usage, but not CSA, can be used to predict V3 loop bnAb sensitivity.
Next, we tested whether coreceptor usage prediction and the estimated CSA between V1 loop and V3 loop bnAbs could help distinguish less susceptible strains that have an N332 site. The CRUSH (coreceptor usage prediction for HIV-1) Web tool was used to predict receptor utilization among all CATNAP database N332-containing Envs with neutralization data against either PGT121 or 10-1074 (21). This algorithm more accurately predicts CXCR4 usage based on V3 loop sequence than WebPSSM or Geno2Pheno (30, 31). Previous clinical trials have used an IC50 below 2 μg/ml as an inclusion criterion for V3 loop bnAb-based therapy, and thus, Envs were classified as sensitive or resistant based on this criterion (2, 3). CRUSH and V1 CSA yielded median areas under the receiver operating curve (ROC) of 0.68 (95% confidence interval [CI], 0.61 to 0.75; P < 0.0001) and 0.55 (95% CI, 0.48 to 0.61; P = 0.14), respectively, for PGT121 (n = 338) (Fig. 7E). For 10-1074, there was also a statistically significant area under the ROC for CRUSH (median, 0.66; 95% CI, 0.57 to 0.75; P = 0.0006), but not for the V1 CSA (median, 0.55; 95% CI, 0.46 to 0.65; P = 0.25) (n = 289) (Fig. 7F).
For both PGT121 and 10-1074, 175 CATNAP N332-positive Envs were randomly selected as a training set to determine a CRUSH cutoff that would achieve a minimum 90% specificity for predicting an IC50 greater than 2 μg/ml. The remaining 163 and 114 N332-containing Envs were used as a test set for PGT121 and 10-1074, respectively. A CRUSH cutoff of 0.16 had 93.0% (95% CI, 89% to 96%) and 91% (95% CI, 87% to 94%) specificity for PGT121 and 10-1074, respectively, against the entire CATNAP data set. The positive predictive value (PPV) (percentage of Envs with IC50 values of ≥2 μg/ml relative to the total number of strains with the characteristics of interest) was 65% for PGT121 but only 33% for 10-1074 (Fig. 7G and H). The smaller number of CATNAP N332-positive Envs with a CRUSH value greater than 0.16 with available IC50 data against 10-1074 (n = 33) than against PGT121 (n = 48) potentially accounts for this difference. The portion of N332-positive Env variants with a CRUSH score greater than 0.16 had between 2- and 3-fold greater likelihood of having PGT121 (P < 0.00001) and 10-1074 (P = 0.007) IC50s of more than 2 μg/ml than strains with predicted glycosylation at amino acid position 332 but a CRUSH score of less than 0.16 (Fig. 7G and H). Thus, N332-containing Envs with a CRUSH score above 0.16 are much more likely to be resistant to V3 loop-directed bnAbs than either all N332-positive strains or those with a score below 0.16. Variants with a CRUSH score below 0.16, however, do not always have an IC50 of less than 2 μg/ml.
The CRUSH score and predicted 10-1074 CSA may have clinical significance.
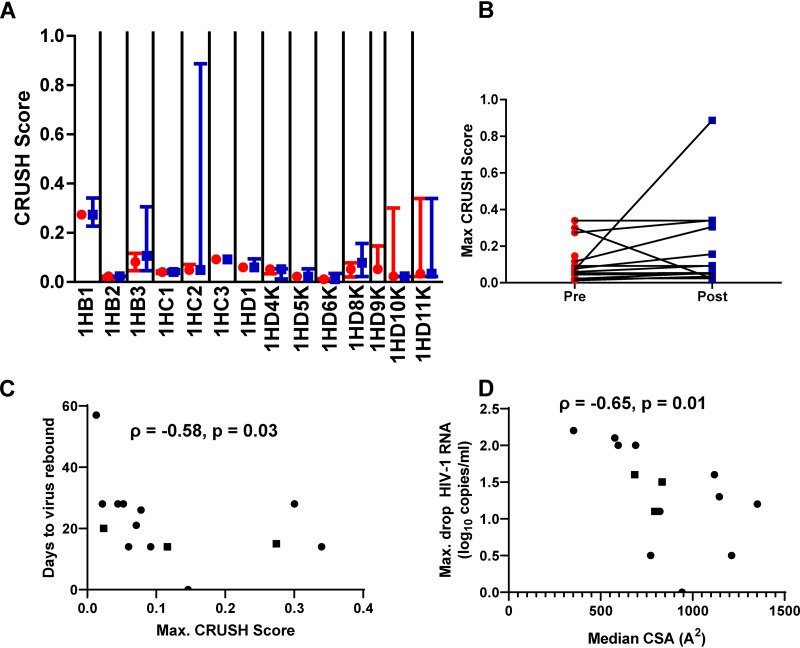
In a 10-1074 monotherapy clinical trial, three individuals (1HB1, 1HD10K, and 1HD11K) with pretreatment Envs with predicted CRUSH scores greater than 0.16 had drops in plasma viremia similar to those of the other subjects (Fig. 8A) (1). There was a statistical trend for the maximum CRUSH score to be higher among the rebound than among the pretreatment sampled Envs (P = 0.06) (Fig. 8B). The CRUSH scores of the preinfection strains were significantly associated with the number of days to viral rebound (P = 0.03) (Fig. 8C). In contrast to the CRUSH scores, there was no significant change in predicted 10-1074 CSAs among pre- and posttreatment Envs (data not shown). The median 10-1074 CSA among the preinfection strains, however, was significantly associated with the maximum decrease in HIV-1 RNA after treatment (P = 0.01) (Fig. 8D). This suggests that individuals with higher CRUSH scores and greater predicted 10-1074 CSAs have faster virus rebounds and smaller plasma virus decreases, respectively, after treatment with 10-1074 monotherapy.
FIG 8.
CRUSH score and predicted CSA are associated with clinical outcomes after 10-1074 monotherapy. (A) Predicted CRUSH scores among pretreatment (red circles) and rebound (blue squares) strains among viremic patients treated with 10-1074 alone. The symbols represent the medians, and the whiskers show the ranges. Individual patients’ IDs are shown below the x axis. (B) Changes in maximum CRUSH scores among pretreatment (red circles) and rebound (blue squares) Envs sampled at different times. (C) Association between preinfection maximum CRUSH scores and the number of days to virus rebound posttreatment. (D) Association between preinfection median 10-1074 CSA and maximum drop in HIV-1 plasma RNA. (C and D) The graphs show Spearman rank correlations. The squares represent the 3 individuals treated with a lower dose of 10-1074.
In contrast to monotherapy, the CRUSH score was not associated with clinical outcomes when individuals received combination treatment with 10-1074 and a CD4 bs bnAb (3BNC117) (2, 3). Three viremic (9342, 9343, and 91C35) and 1 aviremic (9241) individuals had pretreatment variants with CRUSH scores greater than 0.9. Subject 91C35 demonstrated early virus rebound, but the others did not have treatment responses markedly different from those of individuals whose sampled Envs had relatively low CRUSH scores. The maximum CRUSH scores were not higher among the rebound than the pretreatment sampled Envs among the individuals treated with combination bnAbs. The CRUSH scores increased among the rebound compared to pretreatment sampled variants in a significantly higher number of individuals after 10-1074 monotherapy (9 of 13) than after combination treatment (4 of 25) (P = 0.003). Similar to nonhuman primate data, this suggests that V3 loop bnAb monotherapy, but not combination antibody treatment, may promote sequence changes associated with CXCR4 usage (18). While higher CRUSH scores imply that Envs have a greater probability of using the CXCR4 receptor, the pretreatment and rebound strains have not been assessed for coreceptor usage using phenotypic assays.
DISCUSSION
Passive administration of a V3 loop bnAb (10-1074) decreased plasma viremia and delayed virus reemergence in some, but not all, treated individuals (1–3). Presumably, preinfusion virus characteristics impact subsequent treatment efficacy, and it is important to develop methods that predict treatment outcome based on Env properties. In this study, we observed that phenotypically confirmed CXCR4-using variants are less neutralization susceptible to heterologous plasma and to V3 loop N332-dependent bnAbs than R5 variants in some, but not all, cases. As an application of these results, we showed that an algorithm that predicts receptor usage can identify N332-containing variants that are more likely to have decreased susceptibility to V3 loop bnAbs. This coreceptor-based prediction tool, however, cannot reliably identify susceptible strains. We also developed sequence input homology models of envelope-antibody interactions to show that in some cases less neutralization-susceptible variants have relatively large estimated contact surfaces between the Env V1 loop and the antibody. Similar to a previous study of one individual, this suggests that variable-loop interference may impact bnAb potency (47). Sequence-dependent homology modeling provides a potential framework for developing sequence-based tests for estimating bnAb sensitivity, which may be important for future planned bnAb clinical trials (48).
Most bnAb clinical trials have not prescreened patients for antibody susceptibility (1–6). Phenotypic screening using culture outgrowth techniques or Env amplification, cloning, and pseudovirus production is both time- and labor-intensive. Importantly, phenotypic screening has low sensitivity because the methods sample a relatively small proportion of the circulating Envs, and they may miss minor strains that are less susceptible to the bnAb under consideration (49). Although sequence signatures associated with bnAb sensitivity have been defined, it remains uncertain if they can be used in screening tests (13). It is generally agreed that patients with variants lacking a predicted glycan at the Env 332 site would be excluded from V3 loop bnAb therapy. In the CATNAP database, however, a significant proportion of the variants with the N332 site have IC50s above 2 μg/ml. Thus, the presence of the N332 glycan has sensitivities of only around 64% and 78% for PGT121 and 10-1074, respectively. We show that N332-positive variants with estimated CRUSH scores of >0.16 have statistically significant 2- to 3-fold higher likelihood of having IC50s greater than 2 μg/ml than the remaining N332-positive Envs. This CRUSH test, however, had relatively low PPV, because only a small proportion of N332-positive strains in the CATNAP database had CRUSH scores greater than 0.16. The CATNAP database may not be representative of the variants present in patients, because less than 10% of the variants were either phenotypically confirmed or predicted by sequence analysis to use the CXCR4 receptor. Natural history studies, however, estimate that often up to 50% of chronically infected individuals harbor CXCR4-using viruses (50, 51).
A small number of patients have been treated with V3 loop bnAbs, especially monotherapy, and thus, it is difficult to know if a CRUSH cutoff such as 0.16 is clinically meaningful (1–3). Furthermore, our ROC analyses aimed to differentiate Envs with IC50s above and below 2 μg/ml, and it remains uncertain if this IC50 cutoff predicts therapeutic efficacy. Interestingly, the posttreatment variants in patients treated with10-1074 monotherapy had a statistical trend toward higher CRUSH scores. This suggests that V3 loop bnAb infusion selects for either preexisting CXCR4-using strains or mutations that eventually lead to CXCR4 usage. Furthermore, preinfection variants’ CRUSH scores and 10-1074 CSAs were significantly associated with the number of days to virus rebound and the maximum decrease in plasma viremia, respectively. In contrast, in vitro 10-1074 susceptibility of pretreatment culture-derived virus failed to predict outcomes (1). This implies that the Env sequence-based characteristics, both the CRUSH score and the predicted 10-1074 CSA, are superior for predicting treatment efficacy than culture-derived virus sensitivity.
Among CXCR4-using Envs, we showed that predicted V3 loop protrusions either clashed with the CCR5 receptor or eliminated important interactions with amino acids in the CCR5 N-terminal region. Compared to prior studies, this provides a novel mechanistic understanding of the loss of CCR5 receptor usage among some exclusively CXCR4-using Envs (41, 52–54). Env antibody homology models predict that these V3 loop protrusions, however, do not directly limit access to the epitopes important for V3 loop-directed bnAb activity. In contrast, we found that the V1 loop potentially sterically hinders bnAb binding. Thus, the structural basis for the inability to use the CCR5 receptor does not account for decreased sensitivity to V3 loop bnAbs. The impact of V1 loop orientation is another potential reason the algorithm based on V3 loop sequence coreceptor prediction had limited accuracy for identifying N332-containing strains resistant to V3 loop-directed bnAbs.
The HIV-1B V3 loop sequence changes that led to the predicted protrusions are similar to those observed among HIV-1C, HIV-1D, and simian-human immunodeficiency virus (SHIV) X4 strains (14, 15, 55). The forces promoting V3 insertions remain unclear. Neutralizing antibody (nAb) selective pressure has been associated with insertions observed in V1 through V4 Env domains (56–58). Strain-specific V3 loop-directed antibodies that bind at the crown or the base of the V3 loop are common in HIV-1-infected individuals (59, 60). BnAbs, such as PGT121 and 10-1074, also interact with residues in and around the tip of the V3 loop, including the GPG crown and amino acids toward the base of the V3 loop, respectively (44). In aggregate, the similarity in the V3 loop insertions among HIV-1B, HIV-1C, and HIV-1D X4 variants suggest that these divergent viruses are independently converging to a similar solution in response to a common selection pressure, likely nAbs. Isolating antibodies from individuals who harbor X4 strains with V3 loop insertions will provide more definitive proof for this notion.
In general, plasma samples displayed a decreased ability to neutralize Envs in the CXCR4-using strains compared to the global reference panel. The global reference Env collection has been proposed as a standardized panel to evaluate neutralization capacity (39). This panel, however, contains no CXCR4-utilizing viruses. Our results argue that CXCR4-using, especially X4, strains should be included in a standardized Env collection for a more accurate assessment of plasma or antibody neutralization breadth and potency. This may not be important for judging the breadth and potency of potential vaccine-generated antibodies, because nearly all infections are initiated by R5 strains (16, 50, 51). This will be important for potential future antibody-based therapeutics, however, because chronically infected individuals often harbor CXCR4-using strains (50, 51).
Homology modeling was also used to gain a structural understanding of the linkage between differential neutralization susceptibilities and coreceptor usage. The modeling suggested that the orientation of the V1 loop plays a role in influencing susceptibility to V3 loop-directed bnAbs. Notably, these findings further confirm that Env V1-V2 loops have a major impact on sensitivity to autologous, heterologous, and bnAbs (13, 47, 56, 61). As nAbs are introduced in the clinical arena, screening tests can use Env sequences to both predict a phenotype of interest, such as receptor usage, and develop homology structures that incorporate CSA and also electrostatic interactions between different amino acid pairs. This may yield even better sequence-based tests for predicting susceptibility to V3 loop and other bnAbs.
MATERIALS AND METHODS
Study design and samples.
Plasma samples were obtained from the AIDS Clinical Trials Group (ACTG) Study A5095, which was a randomized, double-blind trial assessing different ARV regimens that did not include CCR5 inhibitors (28). All the samples evaluated in this study were obtained before ARV therapy. One sample (SC) from a treatment-experienced individual was also available in the laboratory (40). The ACTG samples were previously characterized for coreceptor usage using both bulk V3 loop sequencing and bulk Env phenotypic analysis (29). Neutralization breadths and potencies were compared among some A5095 samples previously classified as R5 or DM based on these two tests. On the other hand, individual Envs were isolated from samples that were predicted to use CXCR4 based on either bulk V3 loop sequencing or phenotypic analysis.
Envelope isolation, virus stock production, cell lines, and antibodies.
Full-length Envs were amplified from each plasma sample using SGA, as described previously (49). Chimeric Envs were produced using an overlapping-PCR strategy. The specific primer sequences and PCR conditions are available upon request. Amplified Envs were incorporated into an HIV-1 NL4-3 backbone using Saccharomyces cerevisiae gap repair homologous recombination, and virus stocks were generated by human epithelial kidney (HEK) 293T cell transfections as described previously (62). Virus stocks were passaged in peripheral blood mononuclear cells (PBMCs) obtained from HIV-1-seronegative donors for a maximum of 7 days. Viral titers were determined using TZM-bl cells, as described previously (49). All the cell lines and antibodies were obtained from the NIH AIDS Reagent Program.
Genotype prediction, coreceptor usage, and sequence analysis.
Each SGA Env coreceptor phenotype was predicted from the V3 loop sequence using either WebPSSM (30) or Geno2Pheno at a false-predication rate of 5% (31). Phenotypic coreceptor usage was determined by infecting TZM-bl cells in the presence or absence of TAK779 and/or AMD3100, as described previously (49). No Env showed replication in the presence of both inhibitors, and this confirmed that the viruses entered cells only by using one or both of the receptors. Env amplified products were cleaned using ExoSap IT (Affymetrix), and sequences were determined using Sanger sequencing (GenBank accession numbers MK094174 to MK095098).
Env CRUSH scores were evaluated for all CATNAP database variants with a detectable IC50 against PGT121 or 10-1074 and a predicted glycan at amino acid 332. CRUSH scores were also estimated for all available pre- and posttreatment Envs from the 10-1074 clinical trials (1–3).
Neutralization assay.
Neutralization sensitivity was tested by assessing infection of TZM-bl cells in the presence or absence of serial dilution of heat-inactivated plasma or bnAb, as described previously (49). The area under the curve was calculated as described previously (32). None of the plasmas or antibodies demonstrated neutralization against pseudovirions with HIV-1 Env deleted and with vesicular stomatitis virus G envelope protein, suggesting there was no nonspecific inhibition.
The primary X4 variants included in the heterologous panel were 1924-H5 (GenBank accession number MK094773), 3248-37 (MK094963), 3576-31 (MK094990), 4102-80 (MK095018), and 1239-8 (MK094449). The primary R5 variants examined as part of the heterologous Envs included 1486-42 (GenBank accession number MK094498), 3026-B7 (MK094843), SC-3 (MK095092), 0229-69 (MK094217), 3131-34 (MK094945), 1874-4 (MK094743), 1045-57 (MK094307), 2327-3 (MK094786), 1069-29 (MK094325), 1233-B11 (MK094354), and 1389-90 (MK094466).
Neutralization against the global reference and the CXCR4-using Env panels was assessed at only one plasma dilution (1:50), and BP scores were calculated as described previously (33). Heat maps were generated using the Los Alamos HIV sequence database heat map tool (https://www.hiv.lanl.gov/). All the heat maps used hierarchical clustering with the Euclidean distance method. The CXCR4-using Env panel comprised SC4 (GenBank accession number MG940939), DM268Y (KF770429), NL4-3 (AF324493), X1924H5 (MK094773), and X210bB7 (MG940933).
Structural modeling and docking.
Models of X4- and R5-utilizing V3 loops were produced using Rosetta software, made available by the Robetta Structural Prediction Server online. Model 1, the best model based on ProQ2 rank, was selected for each V3 loop. Docking of the CCR5 chemokine receptor with R5 and X4 V3 loops was done using ClusPro (42). All superimpositions were done using PyMOL software. The following HIV-1 structures were chosen by the server to predict the indicated V3 loops (PDB ID 5FYK for 4102 R5 and X4, 1239 R5 and X4, and 1233 R5; PDB ID 3J70 for 3576 X4 and 1924 R5 and X4; and PDB ID 5FUU for 3248 X4).
Env homology models were generated using SWISS MODEL, with BG505 SOSIP.664 as the user input template (43). Env homology models were superimposed on the solved crystal structure of BG505 SOSIP with the PGT121 precursor, 3H+109L (PDB ID 5CEZ), or 10-1074 (PDB ID 5T3X) using PyMOL software (Schrodinger LLC; version 2.2.2). The contact surface area was generated using an open source code available at https://pymolwiki.org/index.php/Contact_Surface. The Env V1 loop was designated amino acids 126 to 157 (HXB2 numbering), and the contact surface between this segment and the antibody of interest was estimated. The CSAs were estimated for all Los Alamos database Envs with detectable IC50s against PGT121 or 10-1074 and a predicted glycan at the Env 332 site. The predicted 10-1074 CSA was also estimated for all preinfection and week 4 posttreatment Envs from the 10-1074 monotherapy trial (1). All the trial patient Env sequences were obtained from GenBank. Individuals with preexisting Envs lacking a predicted glycan at amino acid 332 were not included in the analysis.
Statistical analysis.
Comparisons were done among all Los Alamos CATNAP database Envs with previous neutralization data against specific bnAbs. Envs with phenotypically defined CXCR4 usage were compared to those with exclusive CCR5 usage. Detectable versus undetectable neutralization sensitivity was defined based on the presence of an estimated IC50 less than or greater than the highest tested antibody concentration, respectively. Envs with undetectable IC50s were assigned a value of 100 μg/ml for statistical comparisons.
Comparisons between groups containing independent data points or matched samples were done using the Mann-Whitney test and the Wilcoxon matched-pairs test, respectively. Combined HIV-1B and HIV-1C analyses were done by estimating a proportional difference among cocirculating R5 and X4 variants. This proportional difference was assessed for a significant change from a theoretical median of 1 using the one-sample Wilcoxon signed rank test. Frequency differences were examined using a two-sample test of proportions or the Fisher exact test. Associations were estimated using Spearman rank correlations. ROCs were estimated by separating Envs into groups with IC50s greater and less than 2 μg/ml. Statistical analyses were done using GraphPad Prism 5 (version 5). All P values are based on two-sided tests.
Data availability.
The newly determined sequences were submitted to GenBank under accession numbers MK094174 to MK095098.
ACKNOWLEDGMENTS
This study was supported by NIH grant AI122209 (M.S.). L.R. was supported by AI122209-02S (M.S.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank all of the ACTG Study A5095 participants. We thank Laura White and Wenqing Jiang (Providence/Boston Center for AIDS Research biostatistics core) for helpful discussions. We thank Budhi Sagar for technical insights.
We declare we have no competing interests.
REFERENCES
- 1.Caskey M, Schoofs T, Gruell H, Settler A, Karagounis T, Kreider EF, Murrell B, Pfeifer N, Nogueira L, Oliveira TY, Learn GH, Cohen YZ, Lehmann C, Gillor D, Shimeliovich I, Unson-O’Brien C, Weiland D, Robles A, Kummerle T, Wyen C, Levin R, Witmer-Pack M, Eren K, Ignacio C, Kiss S, West AP Jr, Mouquet H, Zingman BS, Gulick RM, Keler T, Bjorkman PJ, Seaman MS, Hahn BH, Fatkenheuer G, Schlesinger SJ, Nussenzweig MC, Klein F. 2017. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat Med 23:185–191. doi: 10.1038/nm.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bar-On Y, Gruell H, Schoofs T, Pai JA, Nogueira L, Butler AL, Millard K, Lehmann C, Suarez I, Oliveira TY, Karagounis T, Cohen YZ, Wyen C, Scholten S, Handl L, Belblidia S, Dizon JP, Vehreschild JJ, Witmer-Pack M, Shimeliovich I, Jain K, Fiddike K, Seaton KE, Yates NL, Horowitz J, Gulick RM, Pfeifer N, Tomaras GD, Seaman MS, Fatkenheuer G, Caskey M, Klein F, Nussenzweig MC. 2018. Safety and antiviral activity of combination HIV-1 broadly neutralizing antibodies in viremic individuals. Nat Med 24:1701–1707. doi: 10.1038/s41591-018-0186-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mendoza P, Gruell H, Nogueira L, Pai JA, Butler AL, Millard K, Lehmann C, Suarez I, Oliveira TY, Lorenzi JCC, Cohen YZ, Wyen C, Kummerle T, Karagounis T, Lu CL, Handl L, Unson-O’Brien C, Patel R, Ruping C, Schlotz M, Witmer-Pack M, Shimeliovich I, Kremer G, Thomas E, Seaton KE, Horowitz J, West AP Jr, Bjorkman PJ, Tomaras GD, Gulick RM, Pfeifer N, Fatkenheuer G, Seaman MS, Klein F, Caskey M, Nussenzweig MC. 2018. Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature 561:479–484. doi: 10.1038/s41586-018-0531-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caskey M, Klein F, Lorenzi JCC, Seaman MS, West AP, Buckley N, Kremer G, Nogueira L, Braunschweig M, Scheid JF, Horwitz JA, Shimeliovich I, Ben-Avraham S, Witmer-Pack M, Platten M, Lehmann C, Burke LA, Hawthorne T, Gorelick RJ, Walker BD, Keler T, Gulick RM, Fätkenheuer G, Schlesinger SJ, Nussenzweig MC. 2015. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522:487. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bar KJ, Sneller MC, Harrison LJ, Justement JS, Overton ET, Petrone ME, Salantes DB, Seamon CA, Scheinfeld B, Kwan RW, Learn GH, Proschan MA, Kreider EF, Blazkova J, Bardsley M, Refsland EW, Messer M, Clarridge KE, Tustin NB, Madden PJ, Oden K, O'Dell SJ, Jarocki B, Shiakolas AR, Tressler RL, Doria-Rose NA, Bailer RT, Ledgerwood JE, Capparelli EV, Lynch RM, Graham BS, Moir S, Koup RA, Mascola JR, Hoxie JA, Fauci AS, Tebas P, Chun T-W. 2016. Effect of HIV antibody VRC01 on viral rebound after treatment interruption. N Engl J Med 375:2037–2050. doi: 10.1056/NEJMoa1608243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch RM, Boritz E, Coates EE, DeZure A, Madden P, Costner P, Enama ME, Plummer S, Holman L, Hendel CS, Gordon I, Casazza J, Conan-Cibotti M, Migueles SA, Tressler R, Bailer RT, McDermott A, Narpala S, O'Dell S, Wolf G, Lifson JD, Freemire BA, Gorelick RJ, Pandey JP, Mohan S, Chomont N, Fromentin R, Chun T-W, Fauci AS, Schwartz RM, Koup RA, Douek DC, Hu Z, Capparelli E, Graham BS, Mascola JR, Ledgerwood JE. 2015. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci Transl Med 7:319ra206. doi: 10.1126/scitranslmed.aad5752. [DOI] [PubMed] [Google Scholar]
- 7.Schoofs T, Klein F, Braunschweig M, Kreider EF, Feldmann A, Nogueira L, Oliveira T, Lorenzi JC, Parrish EH, Learn GH, West AP Jr, Bjorkman PJ, Schlesinger SJ, Seaman MS, Czartoski J, McElrath MJ, Pfeifer N, Hahn BH, Caskey M, Nussenzweig MC. 2016. HIV-1 therapy with monoclonal antibody 3BNC117 elicits host immune responses against HIV-1. Science 352:997–1001. doi: 10.1126/science.aaf0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunthard HF, Calvez V, Paredes R, Pillay D, Shafer RW, Wensing AM, Jacobsen DM, Richman DD. 2019. Human immunodeficiency virus drug resistance: 2018 recommendations of the International Antiviral Society-USA panel. Clin Infect Dis 68:177–187. doi: 10.1093/cid/ciy463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Julien JP, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, Katpally U, Depetris RS, Stanfield RL, McBride R, Marozsan AJ, Paulson JC, Sanders RW, Moore JP, Burton DR, Poignard P, Ward AB, Wilson IA. 2013. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog 9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PN, Spencer DI, Seaman MS, Schuitemaker H, Feizi T, Nussenzweig MC, Bjorkman PJ. 2012. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A 109:E3268–E3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLellan JS, Pancera M, Carrico C, Gorman J, Julien J-P, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-Ul-Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang G-Y, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang Z-Y, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang L-X, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bricault CA, Yusim K, Seaman MS, Yoon H, Theiler J, Giorgi EE, Wagh K, Theiler M, Hraber P, Macke JP, Kreider EF, Learn GH, Hahn BH, Scheid JF, Kovacs JM, Shields JL, Lavine CL, Ghantous F, Rist M, Bayne MG, Neubauer GH, McMahan K, Peng H, Cheneau C, Jones JJ, Zeng J, Ochsenbauer C, Nkolola JP, Stephenson KE, Chen B, Gnanakaran S, Bonsignori M, Williams LD, Haynes BF, Doria-Rose N, Mascola JR, Montefiori DC, Barouch DH, Korber B. 2019. HIV-1 neutralizing antibody signatures and application to epitope-targeted vaccine design. Cell Host Microbe 26:296. doi: 10.1016/j.chom.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin NH, Becerril C, Giguel F, Novitsky V, Moyo S, Makhema J, Essex M, Lockman S, Kuritzkes DR, Sagar M. 2012. Env sequence determinants in CXCR4-using human immunodeficiency virus type-1 subtype C. Virology 433:296–307. doi: 10.1016/j.virol.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang W, Eshleman SH, Toma J, Fransen S, Stawiski E, Paxinos EE, Whitcomb JM, Young AM, Donnell D, Mmiro F, Musoke P, Guay LA, Jackson JB, Parkin NT, Petropoulos CJ. 2007. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: high prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J Virol 81:7885–7893. doi: 10.1128/JVI.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Regoes RR, Bonhoeffer S. 2005. The HIV coreceptor switch: a population dynamical perspective. Trends Microbiol 13:269–277. doi: 10.1016/j.tim.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Lin NH, Gonzalez OA, Registre L, Becerril C, Etemad B, Lu H, Wu X, Lockman S, Essex M, Moyo S, Kuritzkes D, Sagar M. 2016. Humoral immune pressure selects for HIV-1 CXCR4-using variants. eBioMedicine 8:237–247. doi: 10.1016/j.ebiom.2016.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfeifer N, Walter H, Lengauer T. 2014. Association between HIV-1 coreceptor usage and resistance to broadly neutralizing antibodies. J Acquir Immune Defic Syndr 67:107–112. doi: 10.1097/QAI.0000000000000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sok D, Pauthner M, Briney B, Lee JH, Saye-Francisco KL, Hsueh J, Ramos A, Le KM, Jones M, Jardine JG, Bastidas R, Sarkar A, Liang CH, Shivatare SS, Wu CY, Schief WR, Wong CH, Wilson IA, Ward AB, Zhu J, Poignard P, Burton DR. 2016. A prominent site of antibody vulnerability on HIV envelope incorporates a motif associated with CCR5 binding and its camouflaging glycans. Immunity 45:31–45. doi: 10.1016/j.immuni.2016.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaik MM, Peng H, Lu J, Rits-Volloch S, Xu C, Liao M, Chen B. 2019. Structural basis of coreceptor recognition by HIV-1 envelope spike. Nature 565:318–323. doi: 10.1038/s41586-018-0804-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieslich CA, Tamamis P, Guzman YA, Onel M, Floudas CA. 2016. Highly accurate structure-based prediction of HIV-1 coreceptor usage suggests intermolecular interactions driving tropism. PLoS One 11:e0148974. doi: 10.1371/journal.pone.0148974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Registre L, Moreau Y, Ataca ST, Pulukuri S, Henrich TJ, Lin N, Sagar M. 2019. HIV-1 co-receptor usage and variable loop contact impacts V3 loop bnAb susceptibility. bioRxiv 10.1101/568469. [DOI] [PMC free article] [PubMed]
- 23.Trkola A, Ketas T, Kewalramani VN, Endorf F, Binley JM, Katinger H, Robinson J, Littman DR, Moore JP. 1998. Neutralization sensitivity of human immunodeficiency virus type 1 primary isolates to antibodies and CD4-based reagents is independent of coreceptor usage. J Virol 72:1876–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montefiori DC, Collman RG, Fouts TR, Zhou JY, Bilska M, Hoxie JA, Moore JP, Bolognesi DP. 1998. Evidence that antibody-mediated neutralization of human immunodeficiency virus type 1 by sera from infected individuals is independent of coreceptor usage. J Virol 72:1886–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cecilia D, KewalRamani VN, O'Leary J, Volsky B, Nyambi P, Burda S, Xu S, Littman DR, Zolla-Pazner S. 1998. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J Virol 72:6988–6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bunnik EM, Quakkelaar ED, van Nuenen AC, Boeser-Nunnink B, Schuitemaker H. 2007. Increased neutralization sensitivity of recently emerged CXCR4-using human immunodeficiency virus type 1 strains compared to coexisting CCR5-using variants from the same patient. J Virol 81:525–531. doi: 10.1128/JVI.01983-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho SH, Tasca S, Shek L, Li A, Gettie A, Blanchard J, Boden D, Cheng-Mayer C. 2007. Coreceptor switch in R5-tropic simian/human immunodeficiency virus-infected macaques. J Virol 81:8621–8633. doi: 10.1128/JVI.00759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gulick RM, Ribaudo HJ, Shikuma CM, Lustgarten S, Squires KE, Meyer WA III, Acosta EP, Schackman BR, Pilcher CD, Murphy RL, Maher WE, Witt MD, Reichman RC, Snyder S, Klingman KL, Kuritzkes DR. 2004. Triple-nucleoside regimens versus efavirenz-containing regimens for the initial treatment of HIV-1 infection. N Engl J Med 350:1850–1861. doi: 10.1056/NEJMoa031772. [DOI] [PubMed] [Google Scholar]
- 29.Henrich TJ, McLaren PJ, Rao SS, lin nh, hanhauser e, giguel f, gulick rm, ribaudo h, de bakker pi, kuritzkes dr. 2014. genome-wide association study of human immunodeficiency virus (HIV)-1 coreceptor usage in treatment-naive patients from an AIDS Clinical Trials Group study. Open Forum Infect Dis 1:ofu018. doi: 10.1093/ofid/ofu018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen MA, Li FS, van ‘T Wout AB, Nickle DC, Shriner D, He HX, McLaughlin S, Shankarappa R, Margolick JB, Mullins JI. 2003. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J Virol 77:13376–13388. doi: 10.1128/jvi.77.24.13376-13388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lengauer T, Sander O, Sierra S, Thielen A, Kaiser R. 2007. Bioinformatics prediction of HIV coreceptor usage. Nat Biotechnol 25:1407–1410. doi: 10.1038/nbt1371. [DOI] [PubMed] [Google Scholar]
- 32.Yu X, Gilbert PB, Hioe CE, Zolla-Pazner S, Self SG. 2012. Statistical approaches to analyzing HIV-1 neutralizing antibody assay data. Stat Biopharm Res 4:1–13. doi: 10.1080/19466315.2011.633860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghulam-Smith M, Olson A, White LF, Chasela CS, Ellington SR, Kourtis AP, Jamieson DJ, Tegha G, van der Horst CM, Sagar M. 2017. Maternal but not infant anti-HIV-1 neutralizing antibody response associates with enhanced transmission and infant morbidity. mBio 8. doi: 10.1128/mBio.01373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoon H, Macke J, West AP Jr, Foley B, Bjorkman PJ, Korber B, Yusim K. 2015. CATNAP: a tool to compile, analyze and tally neutralizing antibody panels. Nucleic Acids Res 43:W213–W219. doi: 10.1093/nar/gkv404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 36.Richman DD, Wrin T, Little SJ, Petropoulos CJ. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A 100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koot M, Keet IP, Vos AH, de Goede RE, Roos MT, Coutinho RA, Miedema F, Schellekens PT, Tersmette M. 1993. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med 118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 38.Richman DD, Bozzette SA. 1994. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J Infect Dis 169:968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- 39.deCamp A, Hraber P, Bailer RT, Seaman MS, Ochsenbauer C, Kappes J, Gottardo R, Edlefsen P, Self S, Tang H, Greene K, Gao H, Daniell X, Sarzotti-Kelsoe M, Gorny MK, Zolla-Pazner S, LaBranche CC, Mascola JR, Korber BT, Montefiori DC. 2014. Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 88:2489–2507. doi: 10.1128/JVI.02853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pena-Cruz V, Agosto LM, Akiyama H, Olson A, Moreau Y, Larrieux JR, Henderson A, Gummuluru S, Sagar M. 2018. HIV-1 replicates and persists in vaginal epithelial dendritic cells. J Clin Invest 128:3439–3444. doi: 10.1172/JCI98943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamamis P, Floudas CA. 2014. Molecular recognition of CCR5 by an HIV-1 gp120 V3 loop. PLoS One 9:e95767. doi: 10.1371/journal.pone.0095767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, Beglov D, Vajda S. 2017. The ClusPro web server for protein-protein docking. Nat Protoc 12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garces F, Lee JH, de Val N, de la Pena AT, Kong L, Puchades C, Hua Y, Stanfield RL, Burton DR, Moore JP, Sanders RW, Ward AB, Wilson IA. 2015. Affinity maturation of a potent family of HIV antibodies is primarily focused on accommodating or avoiding glycans. Immunity 43:1053–1063. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gristick HB, von Boehmer L, West AP Jr, Schamber M, Gazumyan A, Golijanin J, Seaman MS, Fatkenheuer G, Klein F, Nussenzweig MC, Bjorkman PJ. 2016. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat Struct Mol Biol 23:906–915. doi: 10.1038/nsmb.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vangone A, Bonvin AM. 2015. Contacts-based prediction of binding affinity in protein-protein complexes. Elife 4:e07454. doi: 10.7554/eLife.07454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van den Kerkhof TL, de Taeye SW, Boeser-Nunnink BD, Burton DR, Kootstra NA, Schuitemaker H, Sanders RW, van Gils MJ. 2016. HIV-1 escapes from N332-directed antibody neutralization in an elite neutralizer by envelope glycoprotein elongation and introduction of unusual disulfide bonds. Retrovirology 13:48. doi: 10.1186/s12977-016-0279-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gruell H, Klein F. 2018. Antibody-mediated prevention and treatment of HIV-1 infection. Retrovirology 15:73. doi: 10.1186/s12977-018-0455-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Etemad B, Ghulam-Smith M, Gonzalez O, White LF, Sagar M. 2015. Single genome amplification and standard bulk PCR yield HIV-1 envelope products with similar genotypic and phenotypic characteristics. J Virol Methods 214:46–53. doi: 10.1016/j.jviromet.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brumme ZL, Goodrich J, Mayer HB, Brumme CJ, Henrick BM, Wynhoven B, Asselin JJ, Cheung PK, Hogg RS, Montaner JS, Harrigan PR. 2005. Molecular and clinical epidemiology of CXCR4-using HIV-1 in a large population of antiretroviral-naive individuals. J Infect Dis 192:466–474. doi: 10.1086/431519. [DOI] [PubMed] [Google Scholar]
- 51.Moyle GJ, Wildfire A, Mandalia S, Mayer H, Goodrich J, Whitcomb J, Gazzard BG. 2005. Epidemiology and predictive factors for chemokine receptor use in HIV-1 infection. J Infect Dis 191:866–872. doi: 10.1086/428096. [DOI] [PubMed] [Google Scholar]
- 52.Chandramouli B, Chillemi G, Giombini E, Capobianchi MR, Rozera G, Desideri A. 2013. Structural dynamics of V3 loop with different electrostatics: implications on co-receptor recognition: a molecular dynamics study of HIV gp120. J Biomol Struct Dyn 31:403–413. doi: 10.1080/07391102.2012.703068. [DOI] [PubMed] [Google Scholar]
- 53.Sander O, Sing T, Sommer I, Low AJ, Cheung PK, Harrigan PR, Lengauer T, Domingues FS. 2007. Structural descriptors of gp120 V3 loop for the prediction of HIV-1 coreceptor usage. PLoS Comput Biol 3:e58. doi: 10.1371/journal.pcbi.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bozek K, Lengauer T, Sierra S, Kaiser R, Domingues FS. 2013. Analysis of physicochemical and structural properties determining HIV-1 coreceptor usage. PLoS Comput Biol 9:e1002977. doi: 10.1371/journal.pcbi.1002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tasca S, Ho SH, Cheng-Mayer C. 2008. R5X4 viruses are evolutionary, functional, and antigenic intermediates in the pathway of a simian-human immunodeficiency virus coreceptor switch. J Virol 82:7089–7099. doi: 10.1128/JVI.00570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sagar M, Wu X, Lee S, Overbaugh J. 2006. HIV-1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection and these modifications affect antibody neutralization sensitivity. J Virol 80:9586–9598. doi: 10.1128/JVI.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rong R, Bibollet-Ruche F, Mulenga J, Allen S, Blackwell JL, Derdeyn CA. 2007. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J Virol 81:1350–1359. doi: 10.1128/JVI.01839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moore PL, Gray ES, Choge IA, Ranchobe N, Mlisana K, Abdool Karim SS, Williamson C, Morris L. 2008. The c3-v4 region is a major target of autologous neutralizing antibodies in human immunodeficiency virus type 1 subtype C infection. J Virol 82:1860–1869. doi: 10.1128/JVI.02187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gorny MK, Xu JY, Karwowska S, Buchbinder A, Zolla-Pazner S. 1993. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J Immunol 150:635–643. [PubMed] [Google Scholar]
- 60.Tang H, Robinson JE, Gnanakaran S, Li M, Rosenberg ES, Perez LG, Haynes BF, Liao HX, Labranche CC, Korber BT, Montefiori DC. 2011. epitopes immediately below the base of the V3 loop of gp120 as targets for the initial autologous neutralizing antibody response in two HIV-1 subtype B-infected individuals. J Virol 85:9286–9299. doi: 10.1128/JVI.02286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pinter A, Honnen WJ, He Y, Gorny MK, Zolla-Pazner S, Kayman SC. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J Virol 78:5205–5215. doi: 10.1128/jvi.78.10.5205-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chatziandreou N, Arauz AB, Freitas I, Nyein PH, Fenton G, Mehta SH, Kirk GD, Sagar M. 2012. Sensitivity changes over the course of infection increases the likelihood of resistance against fusion but not CCR5 receptor blockers. AIDS Res Hum Retroviruses 28:1584–1593. doi: 10.1089/AID.2011.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The newly determined sequences were submitted to GenBank under accession numbers MK094174 to MK095098.