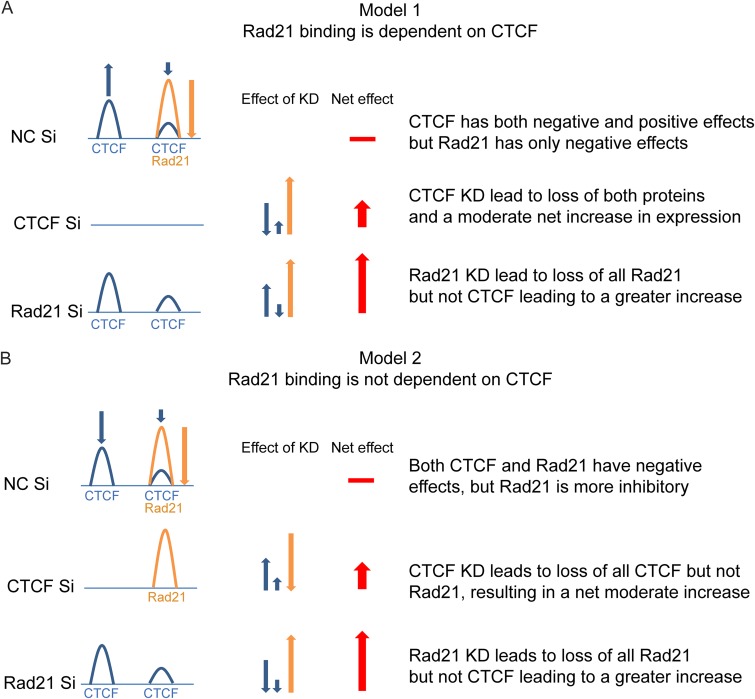
FIG 2.
Models for functional interaction of CTCF and Rad21 on the KSHV genome. (A) Model 1: Rad21 binding is dependent on CTCF binding. Individual or colocalized CTCF (blue) and Rad21 (orange) binding is shown at a regulated gene locus in the KSHV genome. At baseline, CTCF binding can have either positive (blue up arrow) or negative (blue down arrow) effects on transcription, whereas Rad21 has only a negative effect (orange down arrow). Red arrows represent the additive effects of CTCF and/or Rad21 KD on expression at the gene locus. The horizontal red bar depicts the baseline level of gene expression without KD. The predicted effect of CTCF KD is that the loss of CTCF leads to a net decrease in transcription, whereas the concomitant loss of Rad21 leads to a greater net increase in transcription, cumulatively resulting in a moderate increase. KD of Rad21 leads to a loss of Rad21 alone, producing a large transcriptional increase, without affecting the net positive effects of bound CTCF, resulting in a larger net increase than with CTCF KD. (B) Model 2: Rad21 does not depend on CTCF binding. CTCF and Rad21 binding sites are depicted as described above for panel A. CTCF binding, however, is postulated to have only negative effects, similar to Rad21, albeit not as great as those of Rad21. CTCF KD therefore leads to a moderate increase in transcription, and Rad21 KD leads to a larger increase. Neither KD affects the effect of the other protein on transcription.