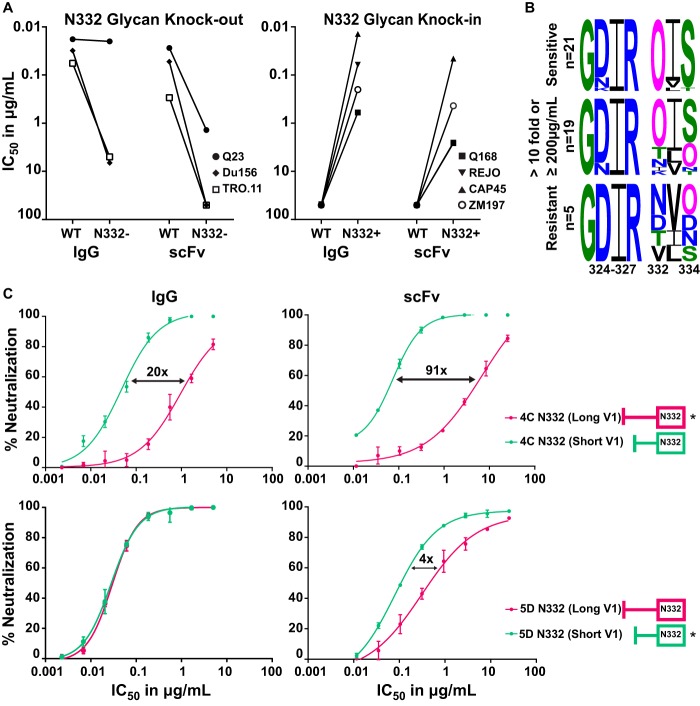
FIG 5.
PGT121 scFv neutralization was dependent on the N332 glycan and the V1 loop length. (A) IC50 of IgG and scFv against 3 sensitive viruses (WT) with their N332 glycan knockouts (N332−) (left) and 4 resistant viruses (WT) with matched N332 glycan knockins (N332+) (right). (B) Noncontiguous amino acid sequences of the V3 region (positions 324 to 327 and 332 to 334, encompassing the GDIR motif and the N332/4 glycans). Viruses are divided into three groups: viruses neutralized equally well by IgG and scFv of PGT121 (top), those with >10-fold difference and/or ≥200 μg/ml for scFv (middle), and viruses resistant to both scFv and IgG (bottom). Glycans [N(X)T/S] are shown as a pink O. (C) Neutralization curves of IgG and scFv (in micrograms per milliliter) against two CAP177 clones, 4C, which has a long V1 (dark pink, star), and 5D, with a short V1 (green, star), and their V1 mutant swaps (no stars). Fold differences between the wild-type and mutant swaps are indicated. scFv is corrected for the molecular weight size difference (multiplied ~5 times).