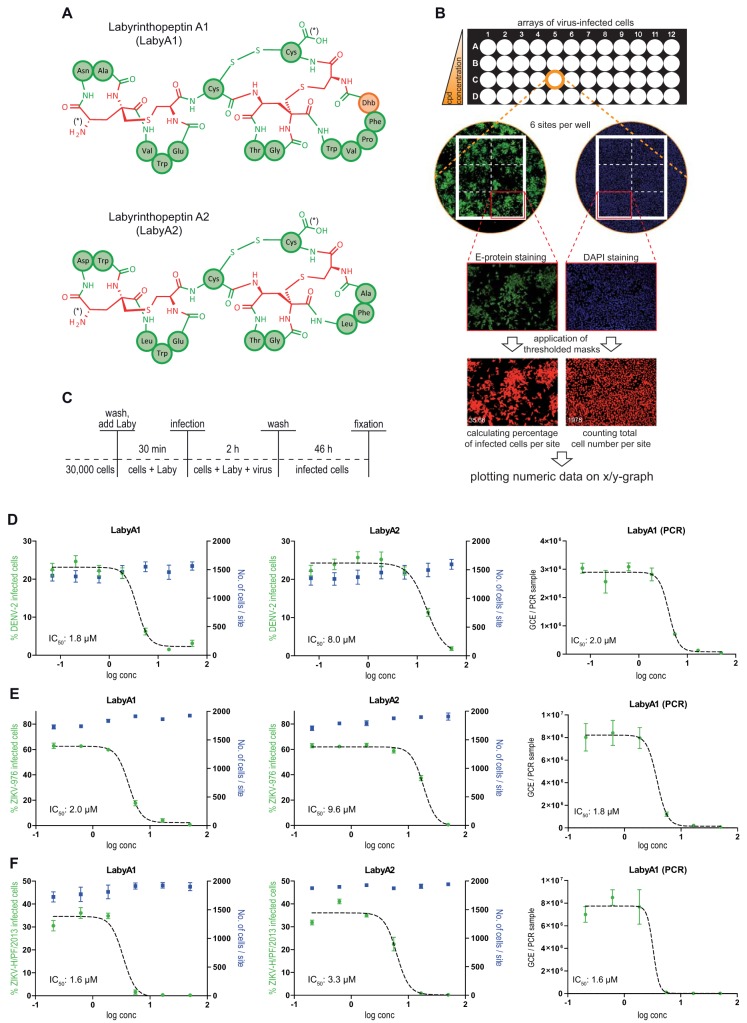
FIG 1.
Labyrinthopeptins efficiently prevent infection of Huh-7 cells with DENV and ZIKV. (A) Schematic primary structures of labyrinthopeptins A1 and A2. The chemical structure of labionin, an αC-quaternary substituted lanthionine, is highlighted in red. Proteinaceous amino acids in three-letter notation are indicated in green. Didehydrobutyrine (Dhb) is indicated in orange. N and C termini are indicated by an asterisk. For further details, see reference 14. (B) Evaluation of viral infection by high-content imaging. Cells were treated with a compound of interest (here LabyA1 and LabyA2) and infected with virus (here ZIKV-H/PF/2013) according to Materials and Methods. Fixed cells were DAPI stained and immunostained for the Zika viral epitope (Alexa Fluor 488). Accordingly processed 96-well plates were analyzed by high-content imaging using an ImageXpressMicro automated microscope (Molecular Devices). Values obtained from the six sites acquired per well were averaged and plotted onto a semilogarithmic X/Y chart. IC50 values were calculated by nonlinear regression using GraphPad Prism. (C) Assay setup for infection of Huh-7 cells with DENV or ZIKV. After fixation, arrayed cells were processed for high-content imaging to assess the number of infected cells (dose-response assay). (D to F) Left and middle panels, dose-response curves obtained for labyrinthopeptin-treated Huh-7 cells infected with DENV-2 (D), ZIKV-976 (E), or ZIKV-H/PF/2013 (F). The total cell number as well as the percentage of virus-infected cells was determined by high-content imaging. Right panels, dose-response curves obtained by RT-PCR from RNA extracted from cell culture supernatants of the same cells. LabyA1 inhibited viral infection of cells more efficiently than LabyA2. Data shown are means of 2 to 5 assays ± SEM. Nonlinear regression was performed with GraphPad Prism.