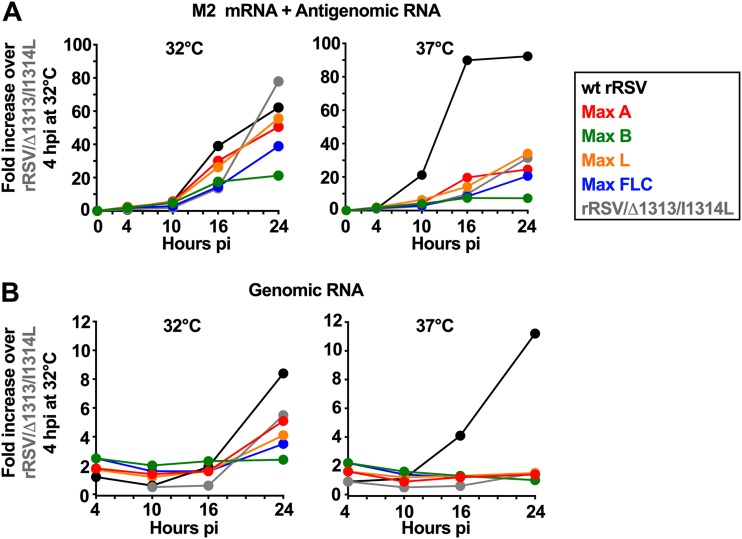
FIG 3.
Effects of CPO on the production of RSV RNAs during single-cycle replication of wt rRSV, rRSV/Δ1313/I1314L, and the CPO rRSVs. Replicate wells of Vero cell monolayers were infected at an MOI of 3 PFU per cell with the indicated viruses or mock infected and then incubated at 32°C or 37°C as indicated. At 4, 10, 16, and 24 hpi, one well per virus was harvested and processed for analysis of cell-associated RNA by strand-specific RT-qPCR, with primers and probe specific to the M2 gene, to detect positive-sense RSV RNA (mRNA and antigenome) (A) or negative-sense RSV RNA (genome) (B). Results were normalized to 18S rRNA. The M2 gene was used for comparison of all viruses because it was the only gene that remained unchanged in the wt and CPO rRSVs. Data are expressed as fold increase over rRSV/Δ1313/I1314 at the 4-h time point.