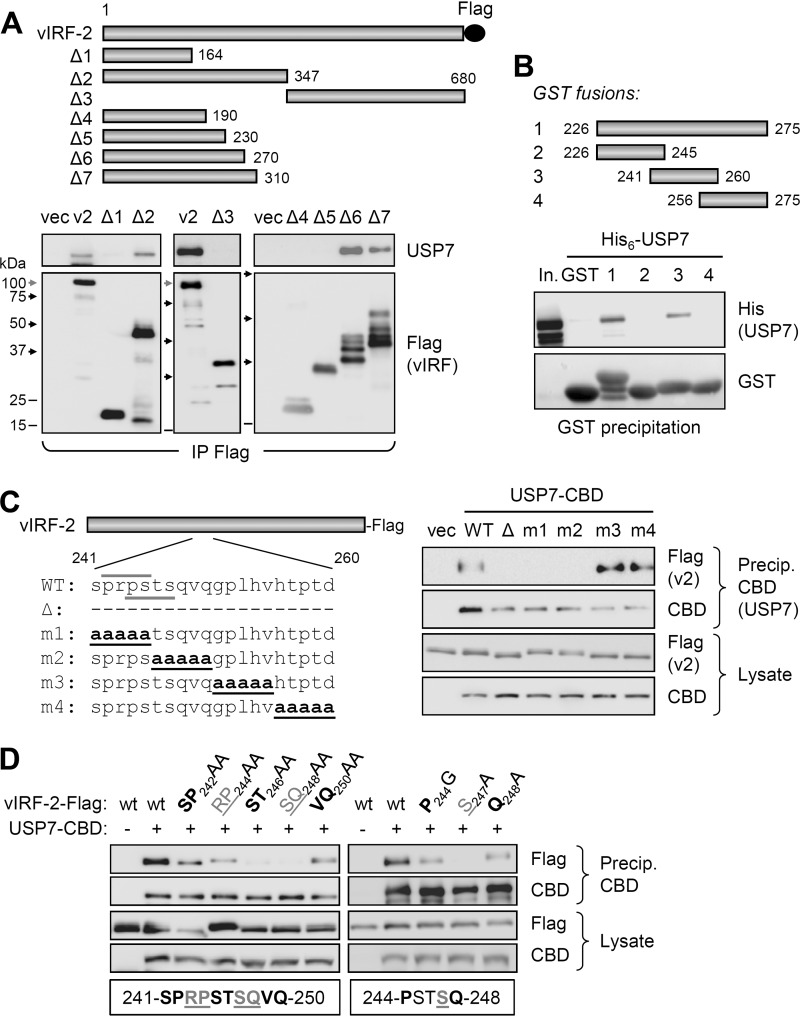
FIG 2.
Identification of vIRF-2 region interacting with USP7. (A) Coprecipitation analysis of (endogenous) USP7 interaction with Flag-tagged vIRF-2 and deleted derivatives (Δ1 to Δ7) expressed in transfected HEK293T cells. The migration positions of marker bands on the different blots are indicated by shaded (100 kDa) and black (75, 50, and 37 kDa) arrows and black lines (25 and 15 kDa). Degradation products (multiple bands) are evident for some of the vIRF-2 deletion variants. (B) In vitro coprecipitation analysis of interaction between recombinant, bacterially derived and purified His6-tagged USP7 and GST-fused vIRF-2 residues 226 to 275 and subfragments 226 to 245, 241 to 260, and 256 to 275. Glutathione bead precipitates were analyzed by USP7 immunoblotting for detection of vIRF-2 fragment-USP7 interactions. In., input His6-USP7. (C) Plasmid vectors expressing the indicated vIRF-2 proteins (left) deleted of (Δ) or mutated (m1 to m4) in the 241- to 260-residue USP7-binding region of vIRF-2 were used in transfection-based coprecipitation assays. Coexpressed CBD-tagged USP7 (isoform 2; NP_001273386.1) was precipitated (Precip.) from transfected cell lysates with chitin beads, and precipitates and lysates were analyzed for USP7-interacting and appropriately expressed vIRF-2 (v2) proteins, respectively. CBD immunoblotting confirmed appropriate affinity precipitation and expression of USP7-CBD. (Left) Over/underlined wild-type (WT) sequences correspond to USP7-binding consensus motifs. (D) Similar analysis of double and single point mutations of vIRF-2 residues 241 to 250. The residues targeted for double and single mutations are indicated below the respective sets of immunoblots of precipitates and lysates from the corresponding transfectants.