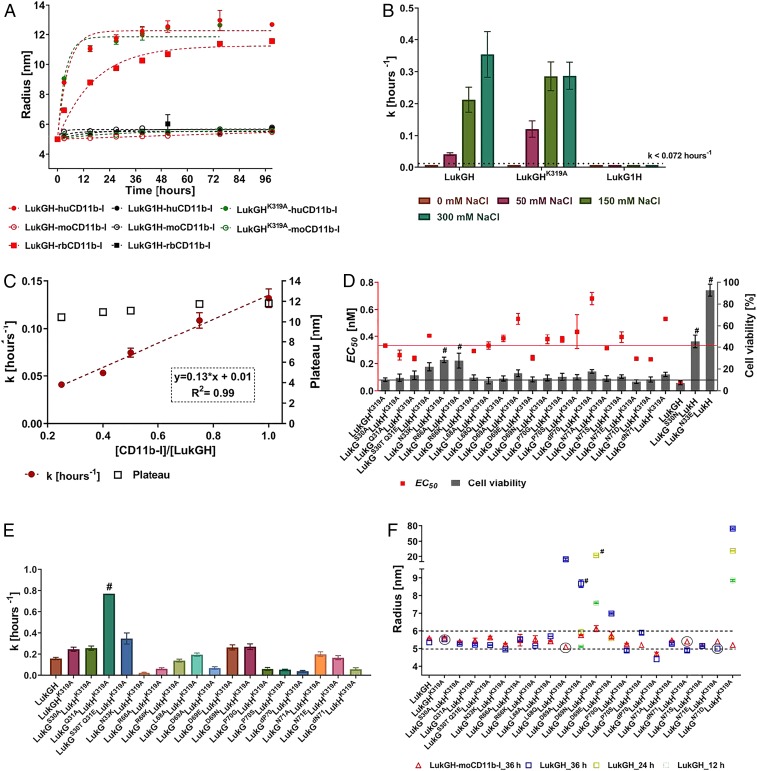
Fig. 3.
Oligomerization of LukGH in solution, binding and activity of LukGH oligomerization variants. (A) Change of LukGH, LukG1H, and LukGHK319A (at 5 mg/mL) plus hu-, rb-, or moCD11b-I (at 2.5 mg/mL) cumulant radius, over time, measured in 25 mM Hepes, pH 7.5, 1 mM MgCl2, 150 mM NaCl (mean of 1 to 2 replicates ±SEM). The dotted lines represent fitting of the data to a one-phase association model with fixed y0 = 5 at x0 = 0 h (GraphPad Prism). (B) Oligomerization rate constant (k) and plateau for LukGH, LukGHK319A, and LukG1H (at 5 mg/mL) plus huCD11bI (2.5 mg/mL) in 25 mM Hepes, pH 7.5, 1 mM MgCl2, 0 to 300 mM NaCl (mean of 1 to 2 replicates ±SEM). Data were fitted as in A giving R2 > 0.93. (C) Oligomerization rate constant (k) of LukGH (4.5 mg/mL) plus increasing amounts of huCD11b-I (2.3 mg/mL) in 25 mM Hepes, pH 7.5, 1 mM MgCl2, 150 mM NaCl (mean of 2 replicates ±SEM). Linear regression fit (GraphPad Prism) is shown in red with equation in Inset. (D) Activity of LukGH mutants toward differentiated HL-60 cells expressed as EC50 and percent cell viability at maximal toxin concentration (100 nM) (mean of 2 independent experiments ±SEM). Red and black line represent EC50 value and percent cell viability of LukGHK319A mutant, respectively. Variants that had limited or no cytotoxicity (could not kill >75% of cells at the highest toxin concentration used) are marked with “#.” (E) Oligomerization rate constant (k) of LukG oligomerization mutants coexpressed with LukHK319A (at 4.5 mg/mL) plus huCD11b-I (2.3 mg/mL) (mean of 2 replicates ±SEM). Data were fitted as in A, in all cases, except for LukGQ31A LukHK319A (#, ambiguous fit), yielding R2 > 0.94. (F) Cumulant radius of LukGH_variant–moCD11b-I complexes (at 4.5 mg/mL for LukGH and 2.3 mg/mL for CD11b-I) and individual LukGH variants at 36 h of incubation in 25 mM Hepes, pH 7.5, 1 mM MgCl2,150 mM NaCl (mean of 1 [circled] or 2 replicates ±SEM). In case the sample shows increased radius at time 36 h, earlier time points are shown (24 and 12 h). Dotted lines represent ±10% change from a 5.5-nm radius. Samples with sum of squares error >10 are marked with “#.”