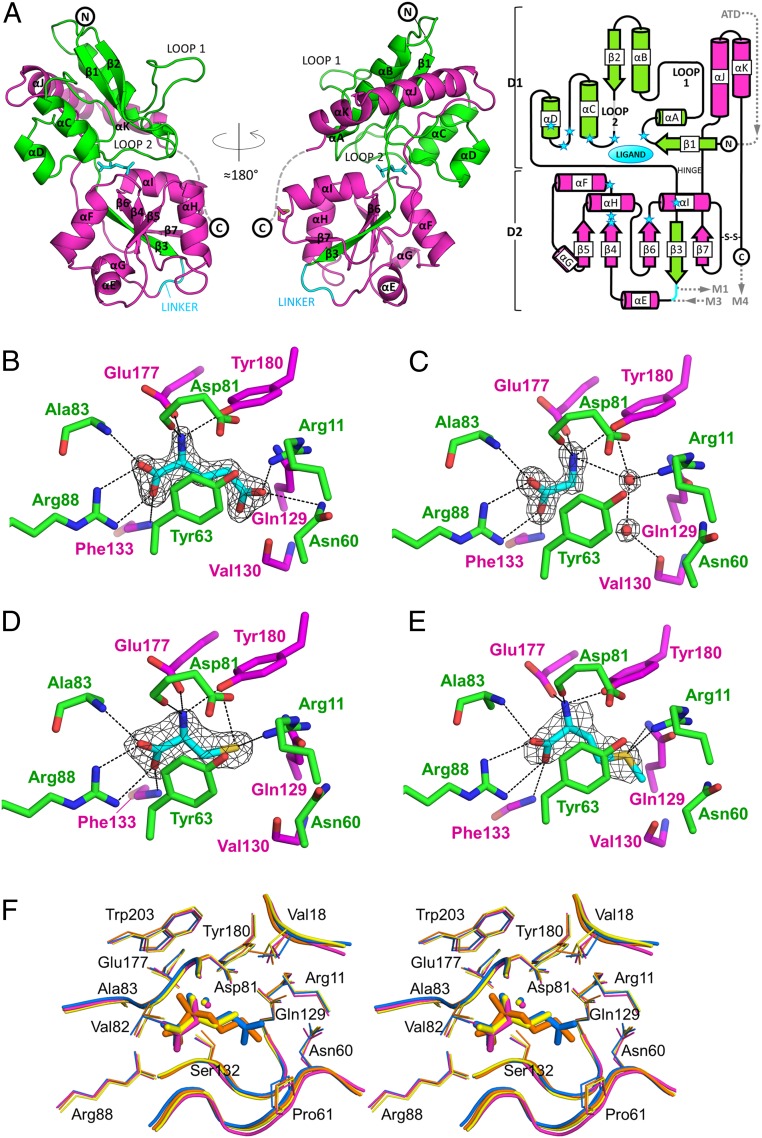
Fig. 2.
Structures of the AtGLR3.3 LBD bound to different ligands. (A) Overall structure of the AtGLR3.3 LBD (+l-Glu) in ribbon representation, colored to highlight the contributions of segments 1 (green) and 2 (magenta) to domains 1 and 2. The linker is colored cyan; l-Glu is in cyan sticks. The C-terminal stretch (dashed) has a defined electron density in 4 out of the 14 protein chains present in the different crystal forms. The structure is oriented in such a way that the N terminus (corresponding to the part of the polypeptide chain right after the ATD domain) is at the top and the linker (replacing the transmembrane segments M1 to M3) is at the bottom. The conventional secondary structure nomenclature used for animal iGluR LBDs has been maintained as reference (including the names loop 1 and loop 2 for the αA-αB and β2-αC loops, respectively). (A, Right) A 2D diagram of the secondary structure of the AtGLR3.3 LBD with the same color code for S1 and S2; cylinders, arrows, and lines represent α-helices, β-strands, and loops, respectively; blue stars indicate the positions of ligand-interacting residues; the position of the ATD and transmembrane domains in the topology of the protein is shown. (B–E) Close-up view of the ligand binding pocket in the crystal structures of the GLR3.3 LBD + l-Glu (B), + Gly (C), + l-Cys (D), and + l-Met (E). The 2|F|o − |F|c electron density omit maps contoured at 1.5 σ are shown for the ligand molecules (cyan sticks) and 2 additional water molecules of the Gly-bound structure (see SI Appendix, Fig. S11 A–D for the corresponding |F|o − |F|c omit maps). The residues or groups of atoms relevant for binding are indicated and represented as sticks, with nitrogen atoms in blue and oxygen atoms in red; protein carbon atoms are either green (if they belong to S1) or magenta (if they belong to S2). Hydrogen bonds are drawn as black dashes; not all interactions are shown for the sake of clarity. (F) Stereoview of the ligand binding site in a superposition of the AtGLR3.3 LBD structures from the 4 datasets. Domain 1 from each structure was superimposed. All side chains (lines) and main chains (tubes) surrounding the ligands are shown, except Tyr63 for clarity. The l-Glu–bound structure is blue, Gly is magenta, l-Cys is yellow, and l-Met is orange. One of the 2 resident water molecules of the pocket is shown; the 2 additional water molecules in the Gly-bound structure that are shown in C are not represented here for clarity.