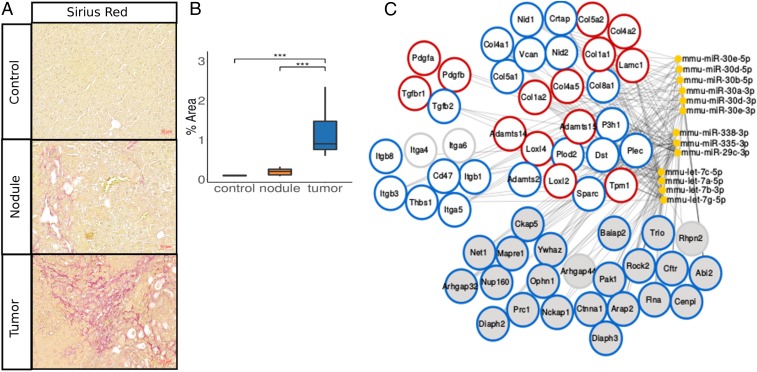
Fig. 1.
A subset of miRNAs predicted to target ECM-linked and fibrosis-associated genes in mHCC. (A) Sirius Red staining of control, nodular, and tumor liver samples isolated from SRF- mice. (Scale bar, 50 m.) (B) Quantification of Sirius Red signal shown in A. (C) Network of miRNA:mRNA pairs, which encompasses predicted miRNA targeting of genes contributing to the ECM-related pathways highlighted in dark gray in SI Appendix, Fig. S1C. Genes are grouped in structural (upper right), signaling (upper left), and remodeling (middle right) components of the ECM, as well as genes related to integrin signaling (middle left) and Rho signaling (bottom). miRNAs mmu-miR-30e-5p, mmu-miR-30d-5p, mmu-miR-338-3p, mmu-miR-335-3p, mmu-miR-29c-3p, mmu-let-7a-5p, mmu-let-7c-5p, and mmu-let-7g-5p, which are predicted to target all ECM-related proteins of the network, were further experimentally characterized in the remainder of this study. Additionally, we chose to further characterize gene expression and predicted miRNA-mediated targeting of a subset of genes, which represent key structural, remodeling, and signaling components of the ECM. We highlighted these genes using red circles. Rims of gene nodes: red, ECM-related genes characterized further in this study and predicted to be targeted by the here-characterized miRNAs; blue, genes not characterized in this study but predicted to be targeted by the here-characterized miRNAs; and gray, genes not characterized in this study and predicted to be targeted by the here-noncharacterized miRNAs. Data are shown as median, first, and third quartiles (“box”) and 95% confidence interval of median (“whiskers”). ***p value 0.001.