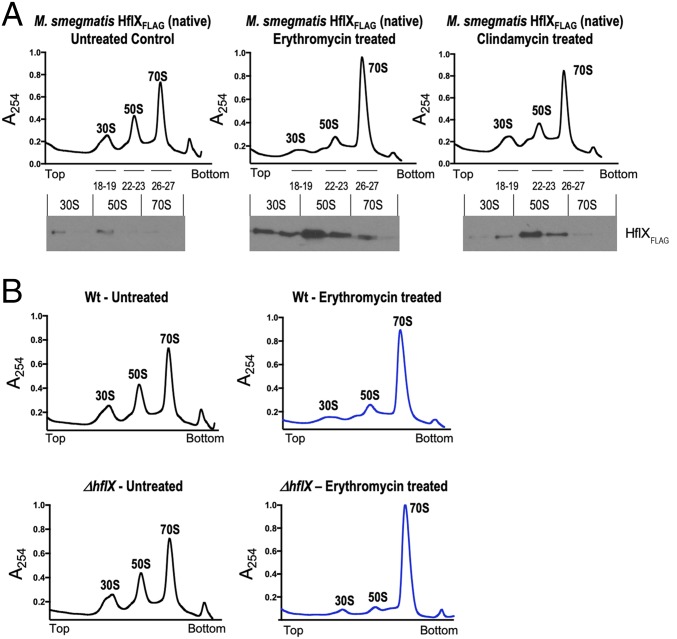
Fig. 3.
(A) In vivo association of Ms-HflX with ribosomes. An M. smegmatis strain in which Ms-HflX was C-terminally tagged with the 3X-FLAG epitope at its native chromosomal location was grown to an OD of 0.7 and treated with either 20 μg/mL erythromycin or 16 μg/mL clindamycin for 1 h. Untreated cells were used as a control. A total of 50 pmoles crude ribosomes isolated from each sample were loaded on a 10-mL, 10% to 40% sucrose gradient, followed by ultracentrifugation in an SW41 rotor. Samples were collected from top to bottom on a Brandel fractionation system, and the distribution of endogenous Ms-HflXFLAG in ribosome fractions was analyzed by immunoblotting, using anti-FLAG antibody. (B) Ribosome profile of ΔMs_hflX compared with wild-type bacteria. Wild-type M. smegmatis and ΔMs-HflX strains were grown to an OD of 0.7 and treated with 20 μg/mL erythromycin for 1 h. Untreated cells were used as a control. Crude ribosomes were prepared from each sample, and equal quantities (50 pmoles) were loaded on 10-mL, 10% to 40% sucrose gradients. After ultracentrifugation, the samples were fractionated using the Brandel Teledyne gradient fractionation system, and results were normalized based on area under the curve (AUC). AUC values for WT-Untreated, WT-ERT treated, ΔhflX-untreated, and ΔhflX-ERT treated were 87.42, 89.66, 86.81, and 87.23, respectively. Polysome profile of erythromycin-treated samples are in blue, and untreated samples are shown in black.