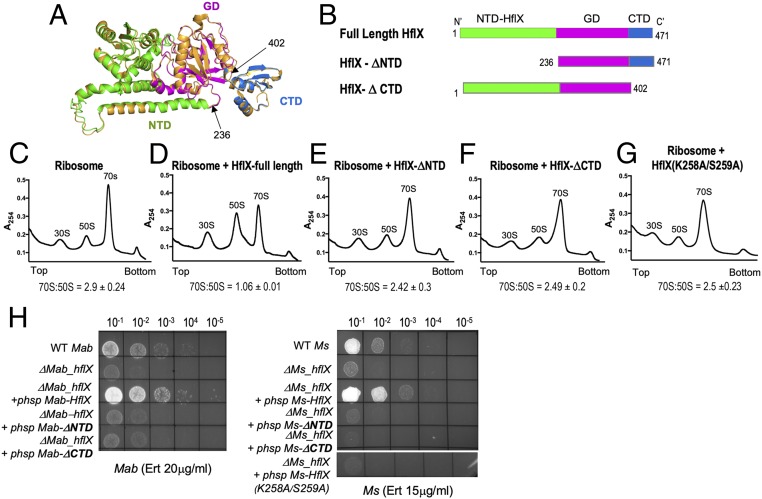
Fig. 4.
Ribosome splitting function of Ms-HflX correlates with its ability to confer antibiotic resistance. (A) A structural model of Ms-HflX guided by the structure of E. coli HflX was obtained using I-TASSER and overlaid on the structure of E. coli HflX (orange) using PyMOL (https://pymol.org/2/). The Ms-HflX model is color coded by domains, as shown on the Right. (B) Location of truncations are indicated. (C–G) Dissociation of 70S ribosomes (0.2 μM) was carried out in the presence of 3.0 μM of either full-length Ms-HflX6his, Ms-HflXΔNTD6his, Ms-HflXΔCTD6his or Ms-HflX(K258A/S259A)6his in HMA-7 buffer containing 1 mM GMP-PNP at 37 °C for 45 min and examined using a 5-mL analytical 10% to 40% SDGC and Brandel gradient fractionation. Percentage AUC was calculated for 70S and 50S peaks, using PeakChart (v. 2.08, Brandel), and expressed as a ratio of 70S:50S. Data represent mean ± SD, n = 3. (H) Wild-type, ΔhflX mutant, and complementing strains containing the respective HflX-ΔNTD, HflX-ΔCTD, and HflX(K258A/S259A) at either the Bxb1 attB site of ΔMs_hflX or the L5 attB site of ΔMab_hflX were assayed for growth on Middlebrook 7H10 containing indicated concentrations of antibiotics. Expression of HflX-ΔNTD and HflX-ΔCTD in the complementing strain was verified using real-time PCR (SI Appendix, Table S2).