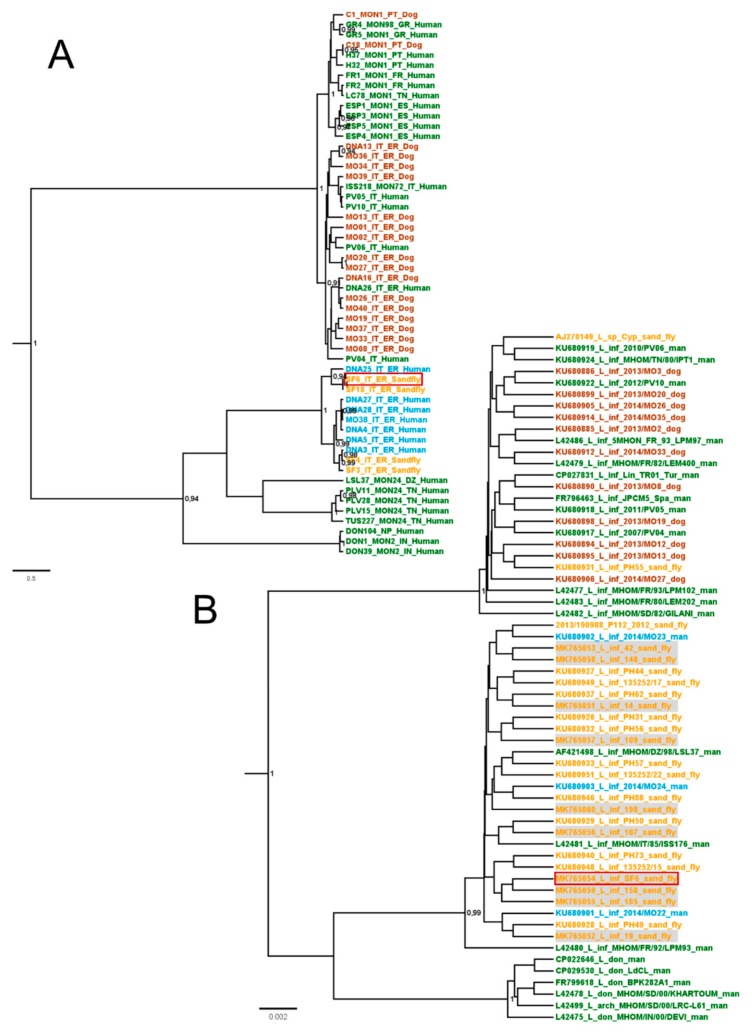
Figure 4.
Bayesian trees obtained by multilocus microsatellite typing (MLMT) (A) and by analysis of the repetitive nuclear region on chromosome 31 (B) of strains of Leishmania from the monitored area and other selected strains. Posterior probabilities over 0.9 showed near the respective node; human strains from the Emilia-Romagna region in azure, human strains obtained from outside the Emilia-Romagna region in green; canine strains in brown, sand fly strains in yellow; sequences obtained in this study are marked in grey, Lein-pw is surrounded by a red square.