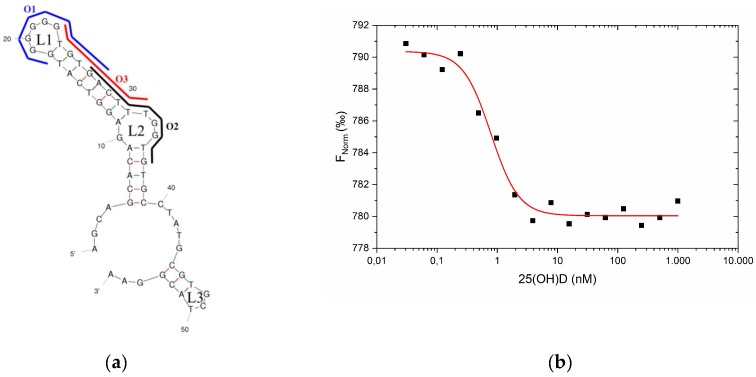
Figure 5.
(a) Illustration of the predicted secondary structure of the VDBA14 aptamer, highlighting binding sites of the complementary designed oligonucleotides using mFold. (b) Oligonucleotide 3 binding to the VDBA14 aptamer at different 25(OH)D concentrations. For the binding curves of oligonucleotide 1 and 2, please see Figure A1. Binding curves were generated by NT.Analysis Software and edited in Origin. The aptamer and oligonucleotide concentrations were kept constant at 10 nM, while the 25(OH)D concentration varied between 1 µM and 0.01 nM.