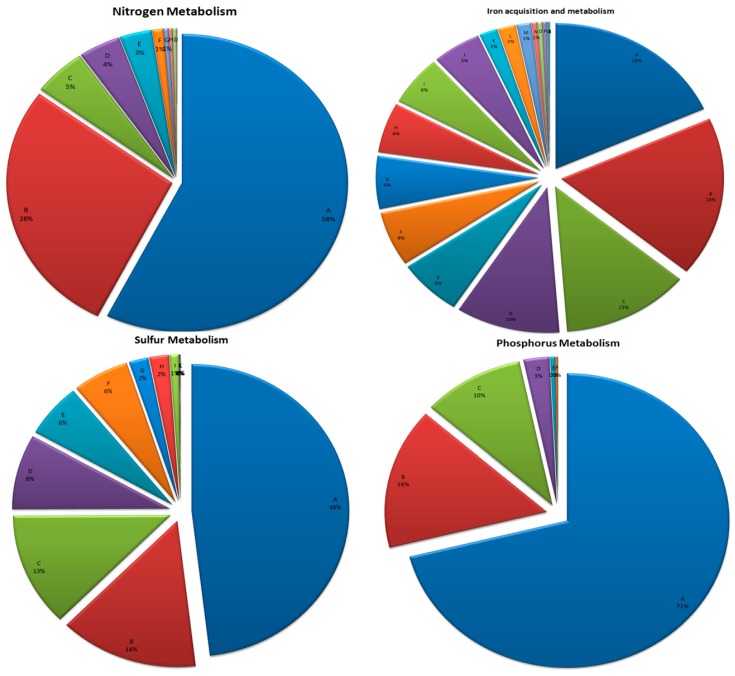
Figure 4.
Assignment of reads in different pathways of the different biological process. Percentage distribution of different pathways in a particular biological process is shown. Abbreviations used are, Nitrogen Metabolism (A: Ammonia assimilation, B: Nitrate and nitrite ammonification, C: Denitrification, D: Nitric oxide synthase, E: Allantoin Utilization, F: Nitrogen fixation, G: Amidase, H: Cyanate hydrolysis, I: Nitrosative stress, J: Nitrilase, K: Dissimilatory nitrite reductase), Sulfur Metabolism (A: Inorganic Sulfur Assimilation, B: Sulfur oxidation, C: Alkanesulfonate assimilation, D: Utilization of glutathione as a sulphur source, E: Thioredoxin-disulfide reductase, F: L-Cystine Uptake and Metabolism, G: Alkanesulfonates Utilization, H: Taurine Utilization, I: Galactosylceramide and Sulfatide metabolism, J: DMSP breakdown, K: Release of Dimethyl Sulfide (DMS) from Dimethylsulfoniopropionate (DMSP), L: Sulfate reduction-associated complexes), Iron acquisition and metabolism (A: Campylobacter Iron Metabolism, B: Siderophore Pyoverdine, C: Transport of Iron, D: Iron acquisition in Vibrio, E: Ferrous iron transporter EfeUOB, low-pH-induced, F: Bacillibactin Siderophore, G: Siderophore assembly kit, H: Encapsulating protein for DyP-type peroxidase and ferritin-like protein oligomers, I: Heme, hemin uptake and utilization systems in GramPositives, J: Heme, hemin uptake and utilization systems in GramNegatives, K: Siderophore Yersiniabactin Biosynthesis, L: ABC-type iron transport system, M: ABC transporter (iron.B12.siderophore.hemin), N: Hemin transport system, O: Siderophore pyochelin, P: Iron(III) dicitrate transport system Fec, Q: Iron Scavenging cluster in Thermus, R: Siderophore Achromobactin, S: Siderophore Enterobactin), Phosphorus Metabolism (A: Phosphate metabolism, B: High affinity phosphate transporter and control of PHO regulon, C: P uptake (cyanobacteria), D: Alkylphosphonate utilization, E: Phosphonate metabolism, F: Phosphoenolpyruvate phosphomutase, G: Phosphate-binding DING proteins).