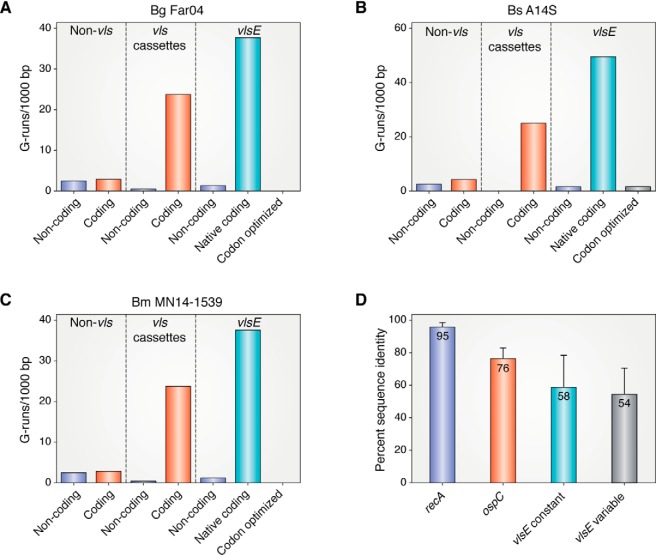
Figure 3.
Distribution of G3+-runs on the linear plasmids carrying the vls loci in Borrelia garinii Far04, Borrelia spielmanii A14S, and Borrelia mayonii MN14-1539 and levels of amino acid sequence identity in RecA, OspC, VlsE constant and VlsE variable regions in Lyme borreliae. A–C, G-runs of 3 nucleotides or more were counted in lp28–9 (CP001316.1), lp28–8 (CP001465.1) and lp28–10 (NZ_CP015805.1) in the three above species, respectively. The distribution of G-runs on both strands of non-vls DNA, in the vls silent cassettes and in the vlsE gene are plotted. The number of G-runs in a codon-optimized vlsE gene generated by reverse translation of the amino acid sequence (https://www.bioinformatics.org/sms2/rev_trans.html) using B. burgdorferi B31 codons (https://www.kazusa.or.jp/codon, species ID: 224326) is also shown. D, Sequence alignments were performed for the 25 available full length RecA sequences that were recovered in a BLAST search (see Table S1 for accession numbers) using DNASTAR MegAlign Pro. The percent identity was determined as the mean of the complete set of pairwise alignments and is shown ± the standard deviation. For OspC and the VlsE variable and N-terminal constant regions the same analysis was performed using sequences from a set of 15 Borrelia strains where both OspC and full-length or near full-length VlsE sequences were available in each strain (see Table S1 for accession numbers). (Please note that the JBC is not responsible for the long term archiving and maintenance of this site or any other third pary hosted site.)