Figure 1.
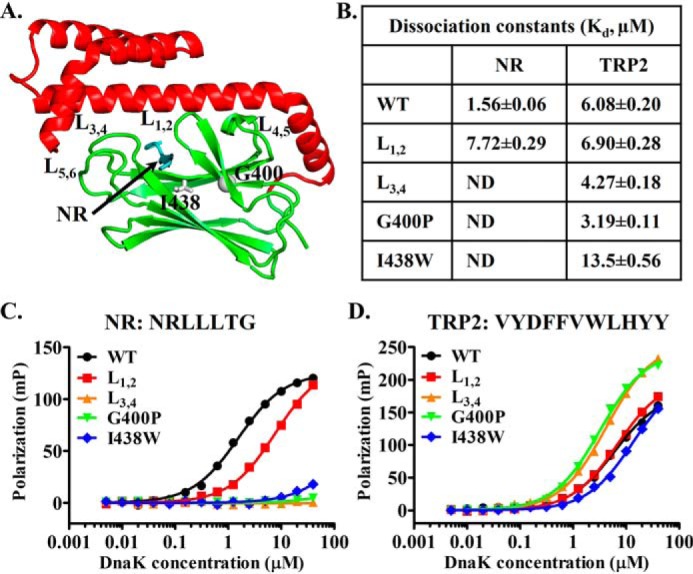
Mutations in the classic peptide substrate–binding site of DnaK have little impact on binding the TRP2 peptide. A, ribbon diagram of the isolated DnaK SBD structure (Protein Data Bank code 1DKX). Ile-438 and Gly-400 are highlighted in gray sticks and ball, respectively. The bound NR peptide is in cyan, and the peptide-binding loops and their supporting loops are labeled. B, the dissociation constants (Kd) of the NR and TRP2 peptides binding from C and D. ND, not detectable. C and D, the binding of DnaK proteins to the NR (C) and TRP2 peptides (D). Serial dilutions of DnaK proteins were incubated with peptides (10 nm), and fluorescence polarizations were recorded after binding reached equilibrium. The sequences of the peptides are labeled on the top of each plot.
