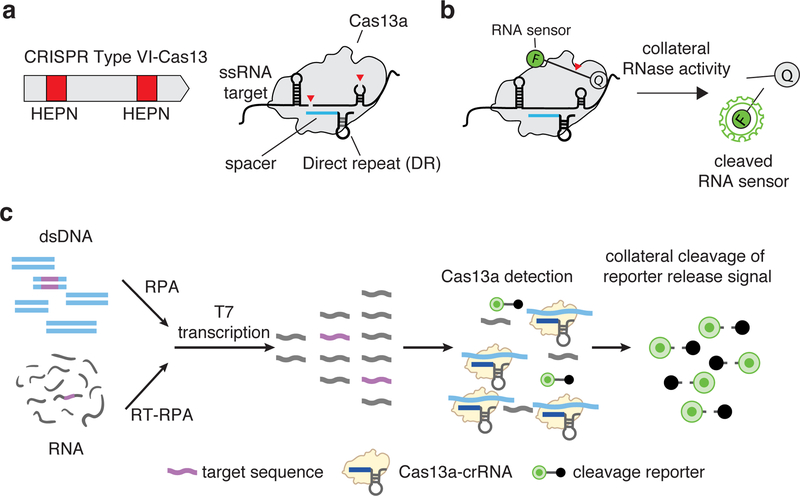
Figure 1: Cas13 complex and collateral activity.
(a) CRISPR-Cas13 RNA targeting complex components. CRISPR-Cas13 enzymes are programmed by a crRNA, which is composed of a direct repeat sequence (DR) flanked by a target complementary spacer sequence (shown in blue). RNA cleavage is mediated by two Higher eukaryotic and prokaryotic nuclease domains (HEPN) domains (shown as red boxes) within a typical Type VI-A Cas13.
(b) Reporter unlocking via CRISPR-Cas13 collateral RNase activity. The CRISPR-Cas13-RNA complex is activated by binding to a complementary target RNA. The activation triggers collateral cleavage of a nonspecific RNA sensor in trans. The fluorescently labeled sensor is quenched when intact and emits fluorescence when cleaved by activated CRISPR-Cas13 complex.
(c) SHERLOCK detection assay Schematic of SHERLOCK assay steps, starting with pre-amplification of either a DNA or RNA target input. Amplified targets are converted to RNA via T7 transcription and are then detected by Cas13:crRNA complexes, which activate and cleave fluorescent RNA sensors. For Cas12 detection, the T7 transcription step is omitted, allowing for direct detection of amplified targets.