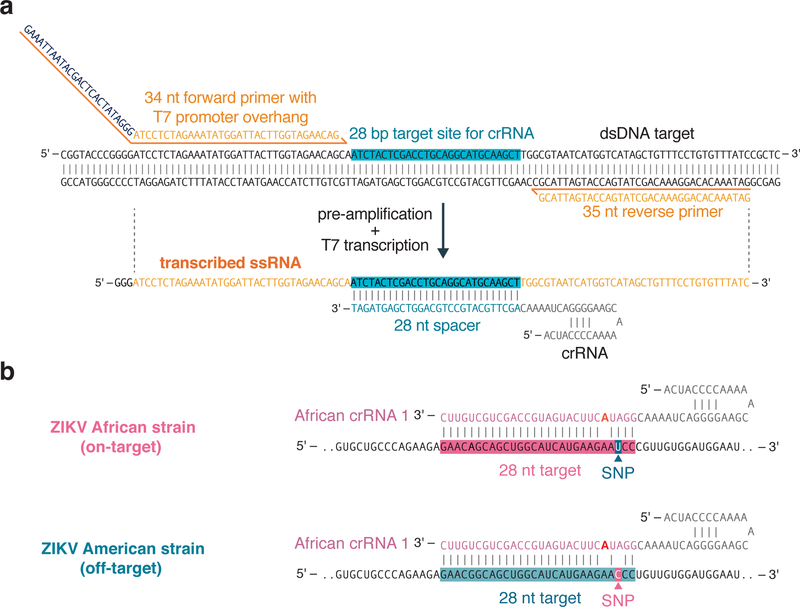
Figure 2: Complete SHERLOCK experimental workflow.
First, LwaCas13a is recombinantly expressed and purified in E. coli (steps 1–28). After crRNA design and in vitro transcription (steps 29–41), sample extraction is performed to yield target nucleic acid (step 42). This sample is then used for RPA-based pre-amplification (steps 43–48) and detection by Cas13 (steps 49–52). Detection can be performed as a single-plex colorimetric lateral flow reaction (step 52 option A), or as fluorescence-based single or multiplex SHERLOCK reactions (step 52 option B). Multiple targets can be detected within the same reaction using CRISPR-Cas13 enzymes with orthogonal cleavage preferences or by combining Cas13 with Cas12 in the same assay.