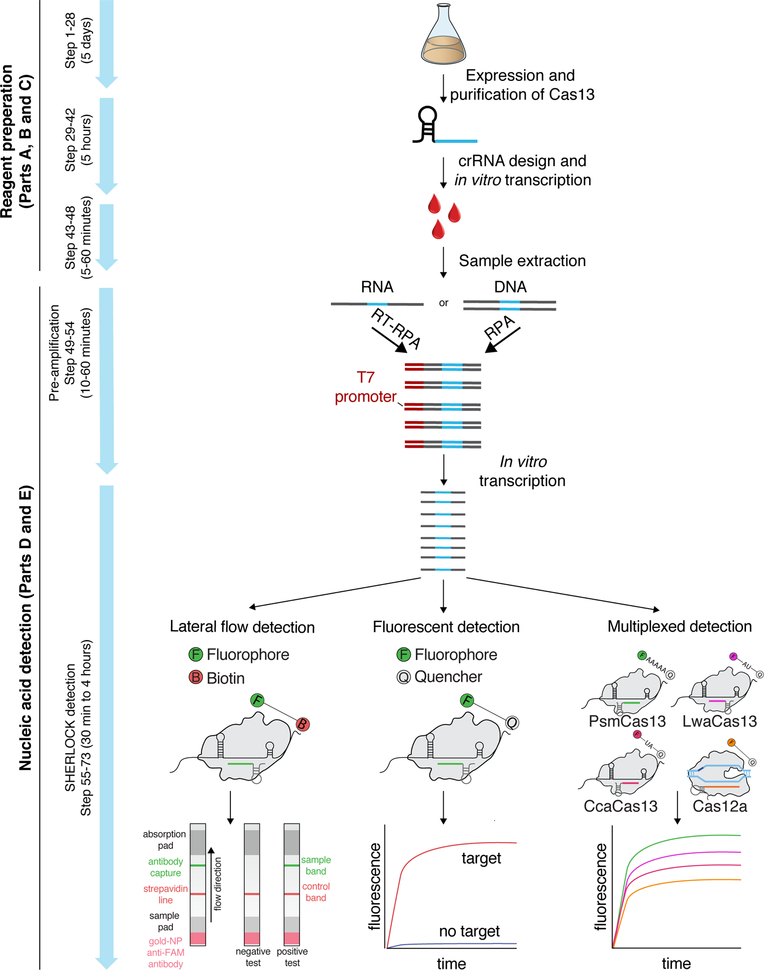
Figure 3: Considerations for primer and crRNA design.
(a) RPA primer and LwaCas13a crRNA design for target detection. Schematic of LwaCas13 crRNA detection of target RNA transcribed from an RNA amplicon. Sequences of the RPA primers and crRNA are shown to highlight how these should be designed relative to the target DNA and transcribed RNA. A T7 promoter should be added to the 5´ end of the forward RPA primer and the crRNA sequence should be the reverse complement of the target site in the transcribed RNA. The direct repeat of the LwaCas13a crRNA is on the 5énd of the spacer sequence.
(b) Synthetic mismatch crRNA design for single-nucleotide specificity. Schematic of crRNA design for sensing a single-nucleotide mismatch difference between African and American strains of the Zika virus. The single-nucleotide polymorphism (SNP) site should be placed in the 3rd position of the spacer sequence and the synthetic mismatch (highlighted in red) should be placed in the 5th position of the spacer sequence.