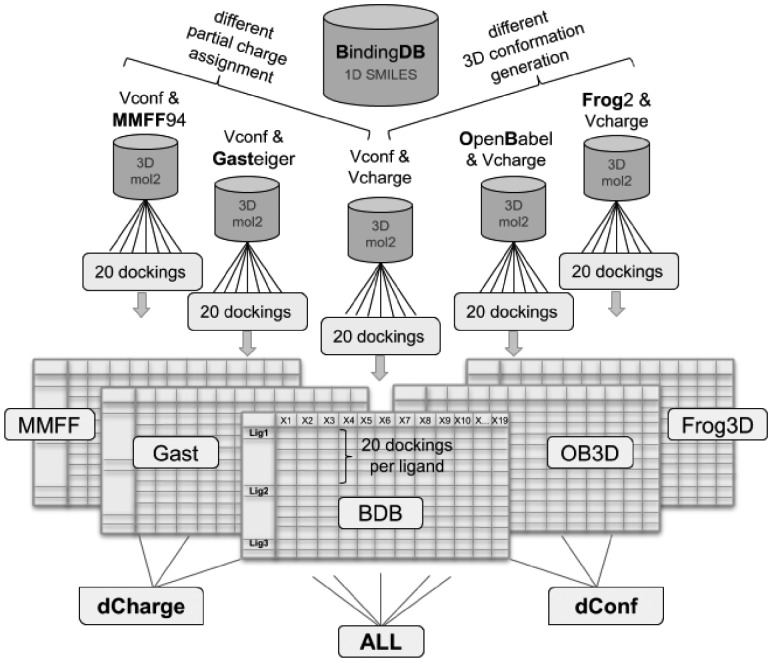
Fig. 1.
Structure-based dataset generation approach. The ligand dataset was extracted from the BindingDB (BDB), which uses VConf for 3D conformation generation and VCharge for charge assignment. Two more partial charge models (MMFF and Gasteiger) and two other 3D conformation generators (OpenBabel and Frog2) were employed to generate a total of five ligand sets. Those were submitted to the @TOME server for docking and complex evaluation. The @TOME output datasets ‘MMFF’, ‘Gast’, ‘BDB’, ‘OB3D’ and ‘Frog3D’ (containing the results of 20 dockings per ligand in different structures) were grouped in three combined datasets, a different charge dataset ‘dCharge’, a different conformation dataset ‘dConf’, and an ‘ALL’ dataset