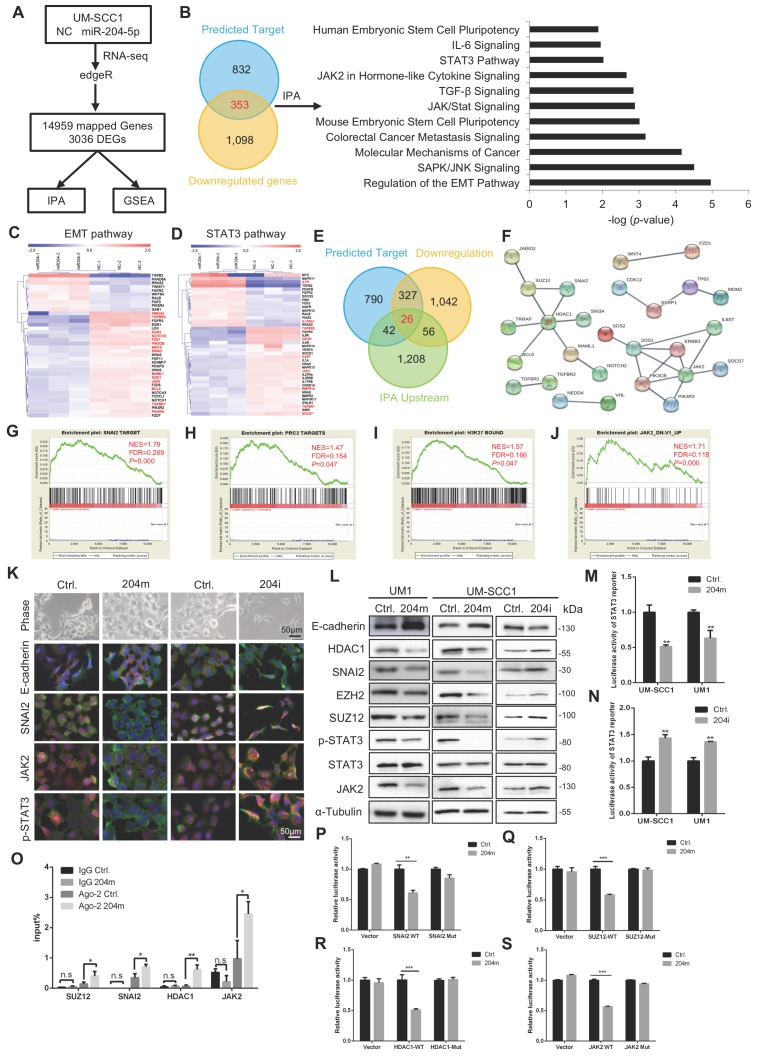
Figure 4.
MiR-204-5p suppresses EMT and STAT3 pathway by targeting SNAI2, SUZ12, HDAC1 and JAK2. (A) Experimental scheme of RNA-Seq in UM-SCC1 cells and subsequent data analysis. (B) IPA showing that miR-204-5p target genes were enriched in metastasis and stemness associated pathway, such as EMT and STAT3 pathway. Heat map of representative EMT (C) or STAT3 pathway (D) related molecules from UM-SCC1 RNA-Seq results. Red font indicates the predicted miR-204-5p targets. (E) Venn diagram showing the predicted miR-204-5p target genes served as upstream regulator based on RNA-seq, IPA and StarBase prediction. (F) PPI prediction among potential targets by using STRING database. GSEA reveals positive enrichment of miR-204-5p-regualted genes in SNAI2 target gene sets (G), PRC2 target gene sets (H), H3K27 bound gene sets (I) and genes up-regulated after knockdown of JAK2 (J). (K) Morphological and Immunofluorescence analysis of cells treated with miR-204-5p mimics and inhibitors. The expression of E-cadherin, SNAI2, JAK2, p-STAT3 (Red) and β-actin (Green) were detected. (L) WB of E-cadherin, HDAC1, SNAI2, EZH2, SUZ12, p-STAT3, STAT3 and JAK2 in HNSCC cells treated with miR-204-5p mimics or inhibitors. STAT3 reporter activities were measured in HNSCC cells treated with miR-204-5p mimics (M) and inhibitors (N). (O) RIP analysis revealed that SUZ12, SNAI2, HDAC1 and JAK2 mRNA were recruited to miRNP complex. Luciferase activity of reporters with wild type or mutant 3'UTRs of SNAI2 (P), SUZ12 (Q), HDAC1 (R) and JAK2 (S) in HNSCC cells co-transfected with indicated oligonucleotides. Data represent mean ± SD. *P < 0.05, **P < 0.01, P < 0.001 by Student's t test.