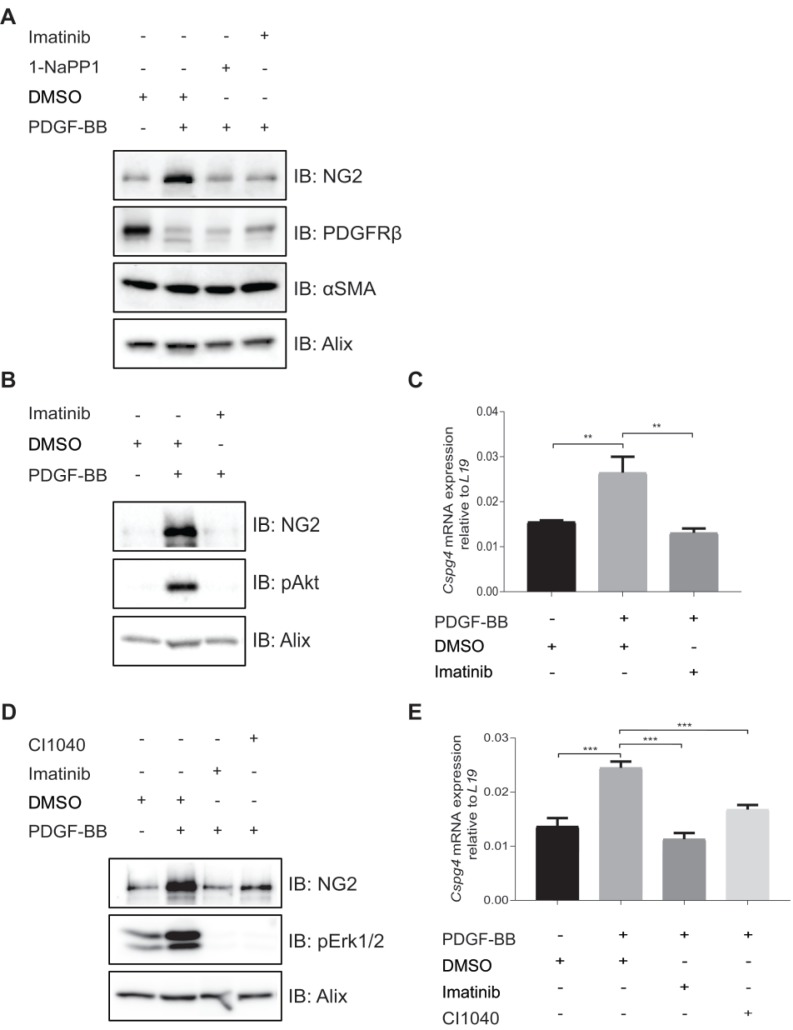
Figure 6.
In vitro inhibition of PDGFRβ affects expression of pericyte markers in ASKA MEFs and 10T1/2 cells. (A) Immunoblotting of pericyte markers in ASKA MEFs, serum-starved and treated with either DMSO, 1-NaPP1 (1 µM) or imatinib (3 µM) in the presence or absence of 20 ng/mL PDGF-BB for 24 h. Total cell lysates were collected and subjected to SDS-PAGE, followed by immunoblotting (IB) for different pericyte markers (NG2, PDGFRβ and α-SMA). IB for Alix was used as a loading control. Representative immunoblots out of three independent experiments, are shown. (B) Immunoblotting for NG2 pericyte marker in 10T1/2 cells, serum-starved and treated with DMSO or imatinib (3 µM) in the presence or absence of 20 ng/ml PDGF-BB for 24 h. Total cell lysates were collected and subjected to SDS-PAGE and the expression of NG2 and phosphorylated pAkt were evaluated by IB. IB for phosphorylated Akt was used to verify receptor stimulation and IB for Alix to verify equal protein loading. Experiment was performed in triplicates and representative immunoblots are presented. (C) To measure Cspg4 (NG2) mRNA expression in 10T1/2 cells, serum-starved and treated with DMSO or imatinib (3 µM) in the presence or absence of 20 ng/ml PDGF-BB for 24 h, we performed quantitative real-time PCR. Error bars indicate standard deviation from triplicate samples. All mRNA expression is relative to L19 ribosomal gene expression. Three independent experiments were performed and statistical analysis was performed by using student t-test. **p<0.01. (D) PDGF-BB induces NG2 expression in a Mek1/2-dependent manner. 10T1/2 cells were serum-starved and treated for 24 h with inhibitors targeting PDGFRβ kinase activity (imatinib, 3 µM) or Mek1/2 (CI-1040, 3 µM) in the presence or absence of 20 ng/mL PDGF-BB. Total cell lysates were collected and the expression of NG2 and phosphorylated pErk1/2 were evaluated by immunoblotting. IB for Alix was used as a loading control and IB for pErk1/2 as a control for the effect of CI1040. Representative immunoblots out of three independent experiments, are shown. (E) To measure Cspg4 mRNA expression, we performed quantitative real-time PCR. Error bars indicate standard deviation from triplicate samples. All mRNA expression is relative to L19 ribosomal gene expression. Three independent experiments were performed and statistical analysis was performed by using student t-test. ***p<0.001.