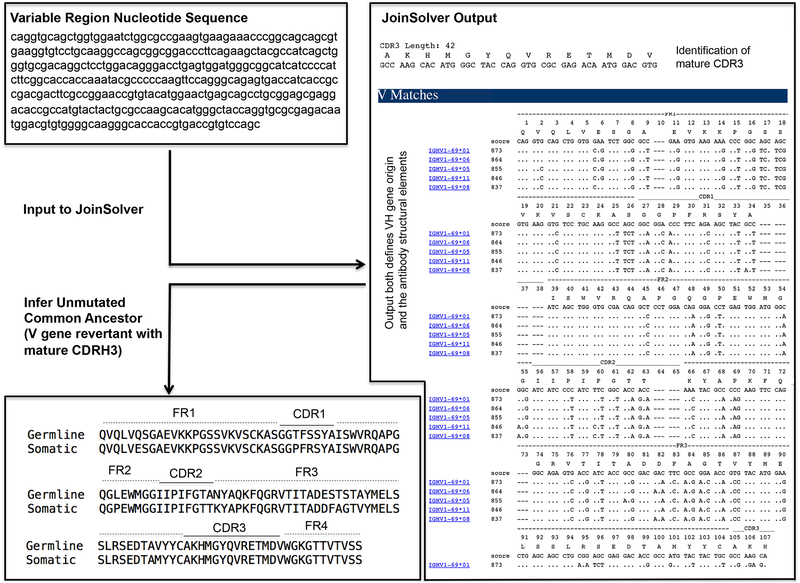
Figure 2.
Inferring a germline reverted antibody. Presented is an example using the heavy chain variable region from CR6261, an IGHV1–69-derived bnAb against influenza virus6,48. The nucleotide sequence for the variable region (FR1, CDR1, FR2, CDR2, FR3, CDR3, FR4) was entered into JoinSolver and analyzed using the Kabat nomenclature41 setting. The two important features of the output are: 1) the highest scoring V gene alignment (=V gene origin); and 2) the position and sequence of the mature CDR3. Information on the D and J genes used to generate the CDR3 are also provided in the output, but they are not displayed or used in this protocol as this junctional information it is often not clear39 and is consequently not standard to germline reversion.