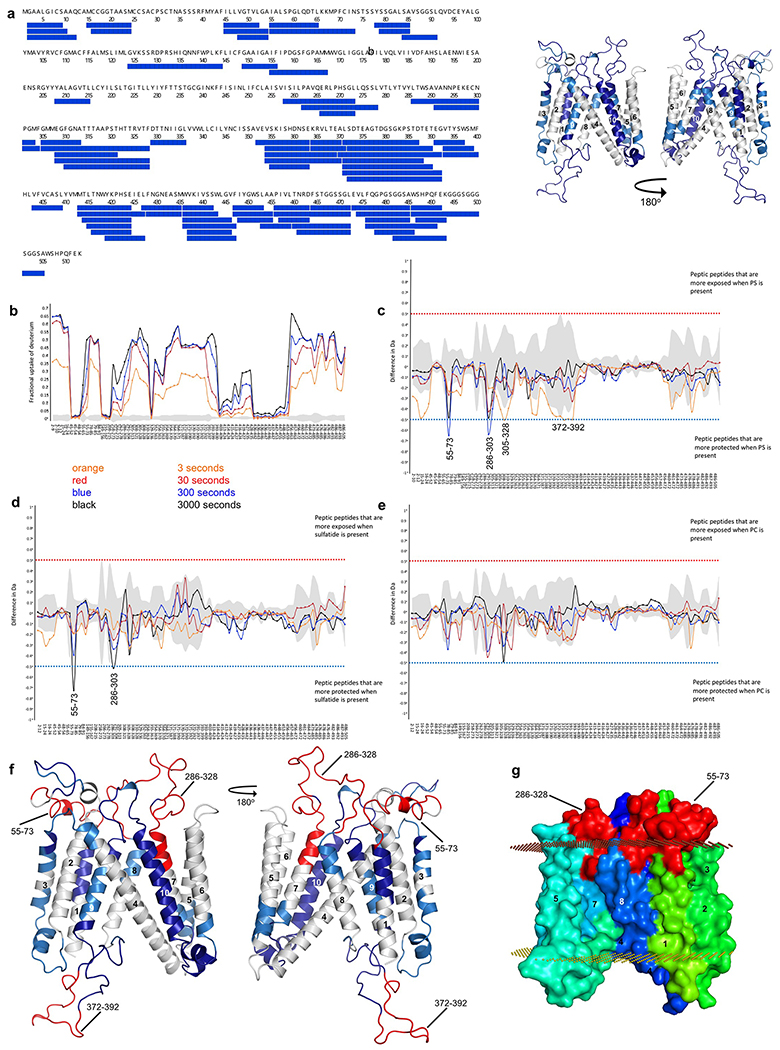
Extended Data Fig. 7. HDX of lipid interactions with DmSERINC.
a, Left: Peptide coverage of DmSERINC monomer for HDX. Right: Structure of DmSERINC (with undefined loops modeled in using SWISS MODEL) with coverage highlighted in blue. b-e, HDX profile of purified monomeric DmSERINC in LMNG micelles prior to (b) or after spiking with exogenous DPPS (c), sulfatides (d), or PC (e) Peptide residue numbers are shown on the x-axis. f, Protected regions determined by HDX mapped onto the DmSERINC structure and highlighted in red (with undefined loops modelled in using SWISS MODEL). g, surface representation of DmSERINC structure colored as in Fig. 1b. with protected regions highlighted in red.