For more than two centuries, biodiversity collections have served as the foundation for scientific investigation of and education about life on Earth (Melber and Abraham 2002, Cook et al. 2014, Funk 2018). The collections that have been assembled in the past and continue to grow today are a cornerstone of our national heritage that have been treated as such since the founding of the United States (e.g., Jefferson 1799, Goode 1901a, 1901b, Meisel 1926). A diverse array of institutions throughout the United States, from museums and botanical gardens to universities and government agencies, maintain our biodiversity collections as part of their research and education missions. Collectively, these institutions and their staff are stewards for at least 1 billion biodiversity specimens that include such diverse objects as dinosaur bones, pressed plants, dried mushrooms, fish preserved in alcohol, pinned insects, articulated skeletons, eggshells, and microscopic pollen grains. In turn, these collections are a premier resource for exploring life, its forms, interactions, and functions, across evolutionary, temporal, and spatial scales (Bebber et al. 2010, Monfils et al. 2017, Schindel and Cook 2018).
Biodiversity collections have historically consisted of physical objects and the infrastructure to support those objects (Bradley et al. 2014). However, the last two decades have witnessed a remarkable wave of digitization that has reshaped the collections paradigm to include digital data and infrastructure (Nelson and Ellis 2018), opening vast new areas for integrative biological research (e.g., a single plant specimen mounted on an herbarium sheet may be analyzed in multitude ways to yield data on flower morphology, DNA for applications from systematic studies to genome sequences, and isotopes for analyses of nitrogen to understand the mechanisms of phenology in relation to nitrogen uptake). In the United States, investment by the federal government through the National Science Foundation's (NSF) Advancing Digitization of Biodiversity Collections (ADBC) program has facilitated the digitization of approximately 62 million US biodiversity specimens since 2011 through 24 thematic collection networks connecting over 700 collections. These networks have helped to develop a collaborative infrastructure connecting specimen data, human resources, research, and education among institutions. The ADBC program has also provided support to iDigBio (the Integrated Digitized Biocollections), which is the central coordinating unit for the digitization effort. The final ADBC grants will be awarded in 2021.
During the last several years, the Biodiversity Collections Network has led an effort to gather input from primary stakeholder communities regarding future directions for collections and their use in research and education. The effort culminated in a workshop held from 30 October through 1 November 2018 at Oak Spring Garden in Upperville, Virginia, during which a strategy was developed to maximize the value of collections for future research and education that builds on and leverages the accomplishments of the ADBC program. The strategy that was informed by stakeholders, refined by workshop participants, and vetted through public comment from scientific community is presented in the present article.
The concept: Extended specimens
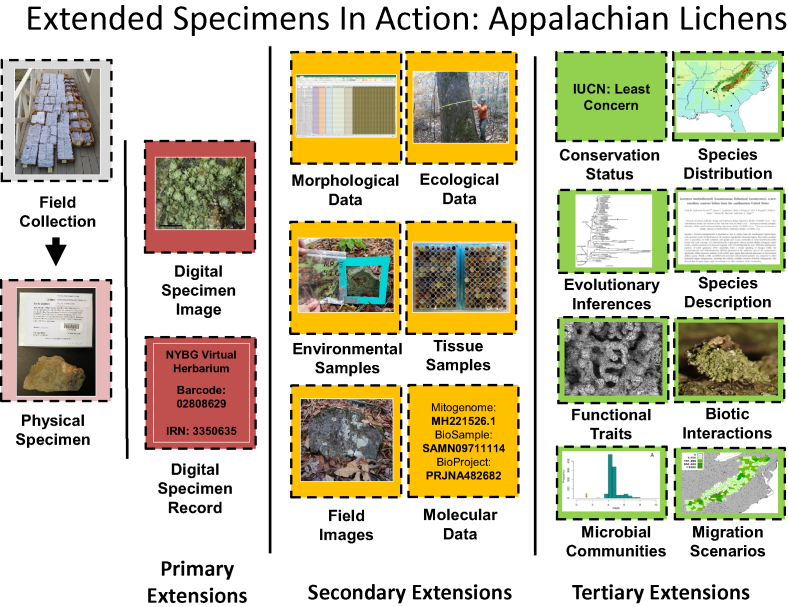
Science, industry, and society rely on physical specimens housed in US biodiversity collections (e.g., Bradley et al. 2014, Tewksbury et al. 2014, Trejo-Salazar et al. 2016, DuBay and Fuldner 2017). Ongoing advances in data generation and analysis have transformed biodiversity collections from physical specimens to dynamic suites of interconnected resources enriched through study over time (Page et al. 2015, Soltis 2017, Nelson and Ellis 2018). The concept of an extended specimen (Webster 2017) conveys the current perspective of the biodiversity specimen as extending beyond the singular physical object to potentially limitless additional physical preparations and digital resources (figure 1; Schindel and Cook 2018).
Figure 1.
Example of an extended specimen generated by the Dimensions of Biodiversity award to study lichen biodiversity gradients in the Southern Appalachian Mountain Biodiversity Hotspot of the eastern United States. The specimen (E. Tripp 6292) was collected in Little River Canyon National Preserve, Alabama, and formally described as Lecanora markjohnstonii by the project team in a paper lead authored by a graduate student from University of Colorado, Boulder. The primary specimen extensions were created and disseminated by The New York Botanical Garden.
The initiative: An extended specimen network
The monumental effort to digitize millions of specimens has resulted in a more accessible and integrated body of information on species occurrence in space and time. Imaging of specimens has provided access to verification and validation of species identification. New and emerging techniques (e.g., CT scanning, isotopes, and AI) are actively unveiling new sources of data and information related to collected specimens. Researchers in biological informatics have provided new methods to analyze and integrate data to address emerging questions. The next step in advancing and enhancing biodiversity collections infrastructure must take into account the rapid development of novel applications for biodiversity data and the growing body of data and resources derived from specimens. Addressing the new hypotheses that researchers may generate, and serving new user communities, will require richer, more complex, and more interconnected networks of information about biodiversity specimens.
The Extended Specimen Network (ESN) is an initiative that would enhance the research potential of specimens, through digitization and links with associated extended data. These extensions will scale from molecules to the ecosphere, and would include genetic, phenotypic, behavioral, and environmental data, as well as biotic interaction networks and new multimedia components (e.g., 2D and 3D specimen images, in situ field images, videos of field conditions). These data reside in disparate databases, have not yet been digitized or made accessible to the scientific community, and are not linked directly to the specimens with which they are associated. Physical specimens are the critical objects that represent the depth and breadth of biodiversity held in US collections, and specimens are the hubs through which these complex data are linked and can be verified and enhanced. The creation of the ESN will leverage and even drive data integration technologies. Once they are integrated and accessible, the extended specimens have the potential to transform the sciences.
The ESN will therefore include the primary physical specimen, all associated physical preparations (e.g., tissue subsamples), and all associated digital data (e.g., micrographs, habit photographs, trait data, genomic data and other molecular markers). The network will involve digitization and extension of existing specimens, and drive the collection of new specimens purposefully collected and accessioned with these extended attributes in mind. The ESN will rely on the development of new data integration mechanisms necessary to link all of the dynamic components together across collections, data types, and existing and evolving databases. These links will help researchers study and better understand the rules that govern how organisms grow, diversify, and interact with one another and how environmental change and human activities may affect those rules.
The ESN is also ideally suited to educating the next generation of data and biodiversity scientists. The combination of object-based learning and digital data will provide a unique gateway to data literacy for the twenty-first century scientist (Petrelli et al. 2013, Hannan et al. 2016). As we look to develop our current and future workforce and foster an informed citizenry, the openly accessible ESN will provide scalable learning opportunities for K–12, undergraduate, graduate, and lifelong learning in data literacy for life sciences.
The ESN will promote discovery
The ESN will stimulate new avenues of investigation, expedite existing ones, and provide an enhanced resource for making science-based policy decisions. By linking physical specimens to the data derived from them (e.g., gene sequences, images, behavior, geographic ranges, and species interactions) and making all of the derived data searchable and easily discoverable, we will have a rich and accessible integrated data source that can be used to advance diverse areas of research. For instance, it will be possible to more fully define and understand the traits that make up organisms, their relationships with each other, and the ecosystems they inhabit. Such information has direct benefits and can inform science relative to fundamental questions that challenge society and the quality of human life. These questions include how crops can be more effectively and efficiently grown in changing climates, how we can sustain and renewably use biological resources in our oceans (Palkovacs et al. 2012), and how zoonotic diseases are transmitted and spread (Samy et al. 2016). In addition to addressing complex questions with broad societal benefits, the ESN will facilitate the as yet incomplete work of documenting and naming the organisms that make up global biodiversity. Machine learning and other novel data science and engineering techniques can help speed identification of museum specimens in the ESN and may aid in recognizing hidden novelties.
Most current portals for digitized specimen data have user interfaces designed for access primarily by biodiversity scientists and collections professionals. To take full advantage of the rich data content and broad relevance of the ESN, the interfaces need to be redesigned to both attract and facilitate use by a broader user base. Certain analyses, data cleaning procedures, and static query parameters, among other features can be added to allow users to interact with the data in more substantive ways. The existing interfaces are set up to retrieve relevant occurrence records from a search on the basis of a taxonomic name, geographic unit, or time period. The ESN will provide an interface that can allow the user to query data in dynamic and discipline specific ways. Future portals, designed for broader uses and broader user bases, could allow for automated queries that include an ability to assess data quality, fitness for use, and information gaps. This would allow users to answer questions such as Do the available data comprise a representative set, or are critical data lacking because key specimens have not been collected or digitized? How many different species, as opposed to different species names, occur in a given region? How many specimens or species in a given query possess a specific trait or suite of traits? Do the organisms of the region occur in populations that are genetically distinct from other populations of the same species? Have unique interactions among organisms been documented in the region of interest?
The ESN will enable seamless data integration, attribution, and use tracking
Central to the success of the ESN is the development and implementation of a system of identifiers and specimen tracking protocols. These will enable dynamic linking of extended specimen components that would otherwise be separated in physical or digital spaces, elucidate relationships between items in disparate collections (tissue:voucher, plant:pollinator, host:parasite, etc.), and facilitate interoperability with data sources outside of our immediate realm (Guralnick et al. 2015). It will also allow collections institutions to follow the use of their specimens and develop metrics for measuring their impact, particularly when used in concert with unique identifiers (e.g., DOIs) assigned to individual data sets. Such metrics permit collections to demonstrate their value, as well as better acquire and manage resources. Currently, collections lack access to the data that demonstrate their full contributions to specimen-based research through citation in publications, vouchering of molecular data (through the National Center for Biotechnology Informatics, NCBI), or products created from direct specimen use (images, CT scans, DNA extracts, etc.).
In addition to providing a discrete and reliable framework for quantifying the impact of biodiversity collections, the new system of tracking will create the potential for cost recovery when specimens are used in commercial enterprise. Use in applied research (e.g., pharmacology, human health, food security) and commercial communities (e.g., pharmaceuticals, agriculture) will further demonstrate the value of collections to additional communities and contribute to the increased sustainability of biodiversity collections.
A further benefit of the new tracking system is that it will also allow biodiversity collections to meet new international standards for documenting the use of specimens and their derived data. An example is the Nagoya Protocol, which is a supplemental agreement to the Convention on Biological Diversity that establishes an international legal framework for access to and methods of sharing benefits of genetic resources. It requires that countries providing specimens define their access procedure and requires users (countries and institutions) to report on and share benefits on their use.
Implementing and sustaining the extended specimen network
Establishing a comprehensive network of extended specimen data that integrates the wealth of biodiversity and expertise held in US collections and associated data repositories will require a monumental effort, comparable to building a new telescope for planetary exploration. However, the focus on digital and human infrastructure, rather than physical infrastructure, makes the magnitude of the effort required to build and sustain such a resource harder to comprehend. Therefore, a major challenge to develop and maintain the ESN will be to establish the unity of the resource, workforce, and scope of effort required to build and maintain it across a network of collections-based institutions.
Creating, growing, and sustaining the ESN requires building and maintaining a network of data providers, a centralized database infrastructure, and an educated workforce. For consistency and sustainability, this requires a central coordinating unit with long term secure funding, which is outside the scope of existing grant programs. Building on the models of the National Center for Ecological Analysis Synthesis, and the National Evolutionary Synthesis Center and the NCBI, a securely funded coordinating center would anchor the ESN and support the data resources. If driven by the stakeholder community, it can serve as a common portal to share established techniques, emerging resources, and common tools for contributing to and sustain the ESN.
We suggest that a distributed ESN platform will be indispensable and that stable funding is required for the ESN to reach its full potential. The envisioned data integration mechanisms would facilitate data integration across major data resources. For example, GenBank (a nondistributed database managed by the NCBI through the National Institutes of Health) data would be integrated within the larger framework of the proposed ESN, thus enhancing the utility of this existing platform by effectively linking genotype, phenotype, and environment. Similarly, data from other new and ambitious efforts such as the Earth BioGenome Project, which aims to sequence, catalog, and characterize the genomes of all of Earth's eukaryotic biodiversity over a period of 10 years (Lewin et al. 2018), and isotopic data from IsoBank would also be integrated with and accessible through the ESN, as would data from new, comprehensive collecting initiatives, such as the NEON Biorepository (e.g., Kao et al. 2012). By integrating the nation's existing critical databases and bioinitiatives, the ESN will greatly enhance the nation's collective biological knowledge, resources, and potential.
Index US biodiversity collections and their holdings
Biological collections comprise the most comprehensive record of life on Earth; their potential will only be fully realized when the data contained within them are revealed and made more accessible for computational analyses. Many of the approximately 1469 US biodiversity collections are not digitized and do not have accurate estimates of the size or taxonomic breadth of their holdings. We need to address this information gap and take stock of the holdings in US biodiversity collections and characterize them in terms of taxonomic, temporal, and geographic emphases, and compile these data into a national collections index. The existing Index Herbariorum (Theirs 2019), an index of plant collections, is a logical model for this resource. An index of all US collections will allow the community to prioritize collections for digitization and facilitate completion tracking for the overall ESN.
Complete and improve existing digitized data
Significant proportions of the existing digitized specimen records are incomplete, with many entered as skeletal records lacking critical data fields (e.g., locality, date, collector). Geocoordinates need entry and verification for the vast majority of digitized specimens. A foundational step in building the ESN will involve completing and standardizing records to maximize their value and interoperability. Development of computational tools that can infer missing values or aid users in completing missing data must be a high priority. Efforts are in place taking advantage of a combination of image analysis (including optical character recognition) and data pattern matching.
Identifying and filling gaps in biodiversity data
Biodiversity specimens have become critical for researchers documenting and investigating environmental change. We must continue to collect specimens and build our collections to inform research into the future. New collections may be especially important in areas that are undergoing rapid change or reduction, such as high-elevation mountain ecosystems, tropical forests, the Atlantic and Gulf coastlines, and the Arctic. Continued collection and integration of new specimens is central to the success of the ESN and as we learn more about how specimens can be used to inform science, we must adjust our sampling protocols to implement a holistic, next-generation approach to the collection of biodiversity specimens. As was discussed by Schindel and Cook (2018), a next-generation approach must be focused on nested sampling that extends beyond the single organism (e.g., a single plant), to include nested sampling of the biotic associates (e.g., soil microbes, epiphytes, endophytes, and parasites spanning from viruses to arthropods and fungi) and data on its environment (e.g., community composition, microclimate, macroclimate, habitat quality). In addition to nested sampling and detailed observations, we need to augment our collecting patterns to conduct targeted sampling to fill existing gaps. Links among nested samples can facilitate new and dynamic research opportunities and transform our capacity to understand life on Earth.
Build and strengthen strategic partnerships
The ESN will enable the expansion of existing partnerships and creation of new strategic engagements. There is a strong need for interdisciplinary development among biology, biological collections, and the computer and data sciences community in order to build, improve, and maintain next-generation collections infrastructure, workforce, and accessibility. We will continue to embrace an openness to collaboration with nontraditional partners in academia and industry that fosters bidirectional benefits.
Build dynamic links with data aggregators
Some of the extended components that need to be linked to existing biodiversity specimens via the ESN are hosted in a growing variety of databases such as the Catalogue of Life checklist, the Biodiversity Heritage Library, the Barcode of Life Data System, the NCBI, and the Encyclopedia of Life Traitbank. Establishing a multidirectional means of data exchange with aggregators is a necessity. Collaboration with programs such as the NSF-funded National Ecological Observatory Network (NEON), the Long Term Ecological Research Network, and Critical Zone Observatories, will ensure that standards and protocols are developed that enable interoperability between collections data and occurrence records both past and present. Furthermore, standardization of extended specimen data driven by the ESN (and the taxonomic expertise across the ESN) will make the data collected by future researchers using these centers computable and therefore accessible to a broad audience of users.
Facilitate data integration across international biodiversity organizations
Participation to the fullest extent possible in the Global Biodiversity Information Facility proposed alliance for biodiversity knowledge (Hobern et al. 2019) will facilitate local work and help align efforts to document, describe, and quantify US biodiversity in relation to other global efforts, including the Atlas of Living Australia and the Distributed System of Scientific Collections, a new European Union program. The ESN also provides the ideal framework within which to identify and address persistent issues in specimen-based data sharing, including the development of standardized taxonomies and ontologies. This could be a strategic goal in conjunction with pursuit of collaboration with data aggregators from Mexico (CONABIO) and Canada (Canadensys) to Brazil (SpeciesLink) to permit the seamless transfer of data needed for large-scale understanding of the breadth of global biodiversity, its distribution and change over time.
The extended specimen network and twenty-first century learners
The ESN has significant transformative potential for scientific research and policy. The ESN can serve as an effective tool to educate and inform broad and diverse audiences about biodiversity and data science. Below we outline the roles that the ESN could have in the realms of formal and informal education.
Formal education
The ESN and collections community has significant potential to engage, educate, and empower the next generation of biodiversity data stewards, researchers, and ESN data users. Biodiversity data and ESN usage, require skills and competencies that align naturally with next-generation science standards outlined for K–12 science curricula (NRC 2013) and the undergraduate biology concepts and competencies included in Vision and Change in Undergraduate Biology Education: A Call to Action (Brewer and Smith 2011). The digital data and specimens central to biodiversity science can be valuable resources and incorporated seamlessly into existing courses, including subject matter in evolution, biodiversity, systematics, taxonomy, and ecology. Specimen-based data make science accessible through the specimen itself, which is tangible, place based, and engaging, as well as through aggregated specimen data that are verifiable, relevant, and a logical gateway to data literacy (Petrelli et al. 2013, Hannan et al. 2016, Monfils et al. 2017). The place-based capacity of collections data combined with the social and societal relevance of biodiversity science can serve a role in creating inclusive, culturally relevant, and socially conscious educational materials that engage a broad and diverse audience in biodiversity science. By defining biodiversity data literacy skills, creating a learning progression that incorporates the ESN and data literacy into formal education, and providing accessible materials with teacher training and educator interfaces that facilitate use in the classroom, we can support an increasingly biodiversity literate society and train the next generation of data literate scientists.
Informal education
As digital resources centered around the ESN expand, so too will informal education opportunities. Indeed, we likely cannot reach the full potential of the ESN without strong involvement of the citizen science community. Many citizen science projects are already structured on monitoring biodiversity: eBird, eButterfly, iNaturalist, and the US National Phenology Network, for example, provide platforms for contributing sightings or recordings of organisms or a particular attribute of an organism. Internet-based projects can involve the public directly in the development and maintenance of the ESN through contributing to collections-based science and databases. Projects such as Notes from Nature (Hill et al. 2012), the Smithsonian Transcription Center, and CitSciScribe are platforms that invite the public to add digital data to images of specimens. Tasks include transcription, morphological measurements, and phenological annotations. Furthermore, the majority of these projects contribute directly to active research projects. Such programs are broadly inclusive and engage participants from a wide range of ages, abilities, and interests, and with minimal start-up costs.
The ESN will advance the NSF’s 10 big ideas
If implemented, the new strategy for biodiversity collections proposed in the present article will provide a powerful scientific tool for the biological sciences. As we outline below, it will also establish a resource that will enable progress toward cross-cutting challenges at the frontiers of science reflected in the 10 big ideas recently identified by the NSF.
Understanding the rules of life
Understanding the rules by which biological and environmental factors influence the wide range of organisms on which humans depend will require diverse data accessed from specimens spanning the tree of life. Biodiversity specimens are a source of genomic, phenomic, and environmental data, the three basic data types required for identifying causal and predictive relationships across these scales. Collections provide these fundamental data, although, as was outlined above, they have not yet been exposed through comprehensive digitization and data linkages. When the vision of the ESN is fully realized, it will be possible to examine key evolutionary traits across spatial and temporal scales, using diverse combinations of data types from the genome to the phenome and beyond. Previously unrecognized patterns may also be discovered using new AI methods, and these could address specific research questions such as how genetic regulators of similar phenotypes evolve in relation to environment.
Harnessing data for twenty-first century science and engineering
The digitization of natural history collections has already contributed to a deluge of data from the nation's scientific facilities. As we broaden the knowledge bank of specimens with additional genotypic, phenotypic, and environmental data, the amount of data will further increase exponentially. The ESN will be a prime example of a cohesive, national scale approach to research data infrastructure through the development and implementation of new tools to integrate data. In addition, specimen-based data provides a unique opportunity to educate students in data science, train a data-enabled workforce, and train future biodiversity data professionals. Collections data, alongside the archived specimens, are an engaging and accessible data source that can provide students an opportunity to experience the entirety of the data pathway while practicing verifiable and testable science.
Midscale infrastructure
Much of the support required for the ESN falls in the gap between what the NSF currently funds as either small or large infrastructure. Specifically, the computer and human infrastructure required for data acquisition, deployment, and training to support the ESN represent midscale infrastructure needs, both in terms of funding level and the scientific research it will support. Although many aspects of the ESN would be characterized as midscale infrastructure, it is important to highlight that support for the ESN coordinating center itself would likely fall within the scope of large infrastructure.
Navigating the new Arctic
The Arctic biota has been minimally sampled over the past two centuries. Arctic biodiversity specimens are stored in relatively few US collections and provide our best baselines for understanding the implications of rapid environmental changes on life (e.g., Bond et al. 2015) in a geopolitical region that is increasingly critical to the global economy and security. Because of the rapid rate of annual warming in the Arctic, we should immediately build international coalitions that will invest in spatially extensive, site intensive natural history collections that will provide the biodiversity infrastructure necessary to critically assess change. Over time, specimens and their associated digitized data, accessed via the ESN, would provide the samples required by diverse technologies (e.g., genomics, isotope ecology), a robust historical context for the proposed network of observational platforms, and a reference for the identification, distribution, behavior, and response of species and their pathogens in the Arctic (Cook et al. 2013, Hoberg et al. 2013).
Growing convergent research at NSF
Enhancement of the roles that collections-derived data can play in understanding and protecting human health, and in education, are key objectives of the new agenda. This could be achieved through the centralized efforts to integrate and share data more effectively and widely. The ESN combines diverse data sources linked to physical biodiversity specimens and, as such, is a physical manifestation of convergent research that will clearly demonstrate the power of this approach.
The future of work at the human technology frontier
The use of biodiversity collections in projects to both refine current techniques in machine learning and to document variation among organisms as exhibited by specimens holds great promise (e.g., Carranza-Rojas et al. 2017, McAllister et al. 2018). The use of these tools will lead to new avenues of data analysis, requiring new skills for researchers and data managers. Collections professionals will find themselves at this frontier. Supporting the adoption of new technology and documenting the experience through training and best practices is a key aim of the strategy presented in the present article. The work carried out by this community may prove transferable to other communities using the tools and workflows developed as part of the ESN.
Enhancing science and engineering through diversity
The ESN requires new generations of taxonomic expertise. The place-based capacity of biodiversity specimens and associated data combined with the social and societal relevance of biodiversity science can serve a role in creating inclusive, culturally relevant, and socially conscious educational materials that engage a broad and diverse audience in biodiversity science. Biodiversity collections institutions have significant potential to engage a broad range of people to create a scientifically literate work force and build the ranks of both the scientific and engineering communities.
Immediate action items to support the ESN
Although a long-term funding structure for the ESN will take time to develop, steps can be taken immediately to initiate the process, including the following:
A robust, comprehensive specimen identifier system should be developed in collaboration with other international data aggregators and providers to enable transparent and uncomplicated integration of biodiversity data with other data sources. The system must also facilitate the attribution of collections’ role in discovery, policy, and promoting transparency for broader issues involving multiple stakeholders (e.g., access and benefit sharing).
An authoritative, comprehensive, and self-updateable index of US collections institutions should be created, similar to the Index Herbariorum for global herbaria, with structured metadata to describe their holdings. This is the first step toward expediting the discovery of undigitized collections and revealing these to the research community.
The digitization of existing material should continue, with a focus on underrepresented taxa (e.g., those in entomology, paleontology, and archeology collections), including data gaps in time, space, and scale, and incorporating of specimens held in small regional and individual researcher collections. These efforts must also include improvement of previously digitized specimen data by imaging specimens, completing skeletal records, and augmenting data with georeferencing.
New protocols must be developed for the collection and dissemination of data-rich samples and nested samples that provide greater context for understanding the biotic and abiotic interactions of organisms, and comprehensive data sets must be created for research and education.
An accessible educator and student interface, baseline analysis tools, and vetted educational materials should be developed to enable integration of biodiversity data in K–12 and undergraduate course work. New tools and resources, combined with training opportunities, will facilitate educator adoption of data-centric educational materials that foster student engagement with digital biodiversity data. Creating an accessible entry into digital biodiversity data will enable training of an informed, digitally fluent workforce.
Broad-scale adoption of core biodiversity data literacy skills and competencies in K–12 and undergraduate curricula should be championed in order to foster a biodiversity savvy future workforce, engage new end users in novel uses of biodiversity data, and sustain and promote careers in advancing collections science, biodiversity research, and data literacy.
Enhanced training of emerging and established professionals for interdisciplinary work in biodiversity, data science, and informatics should be supported. Team Science skills to work collaboratively across disciplines should be emphasized, together with soft skills related to communication, creativity, and critical thinking, because these are paramount to conducting transformative science, communicating research, and cultivating and leveraging relationships with new research and user communities.
Acknowledgments
This material is based on work supported by the National Science Foundation grant no. DBI-1441785. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Author Biographical
James C. Lendemer (jlendemer@nybg.org) is a lichenologist and assistant curator at the New York Botanical Garden, in the Bronx, New York. Barbara M. Theirs, Anna K. Monfils, Jennifer Zaspel, Andrew Bentley, John Bates, David Jennings, Linda S. Ford, and Robert E. Gropp are members of the Biodiversity Collections Network Advisory Committee. BMT is director of the herbarium at The New York Botanical Garden, in the Bronx, New York. AKM is director of the herbarium and a professor at Central Michigan University, in Mt. Pleasant, Michigan. JZ is a research curator and head of zoology at the Milwaukee Public Museum, in Milwaukee, Wisconsin. AB is the collection manager of ichthyology and bioinformatics manager at the University of Kansas Natural History Museum, in Lawrence, Kansas. JB is an associate curator and section head of life sciences in the Integrative Research Center at the Field Museum of Natural History, in Chicago, Illinois. DJ is an iDigBio project manager at the Florida Museum of Natural History, at the University of Florida, in Gainesville, Florida. LSF is the director of collections operations at the Museum of Comparative Zoology, at Harvard University, in Cambridge, Massachusetts. REG is the executive director of the American Institute of Biological Sciences, in Washington, DC. Elizabeth Ellwood is a research fellow at the La Brea Tar Pits and Museum, Natural History Museum of Los Angeles County, in Los Angeles, California. Katherine LeVan is a biorepository scientist for the National Ecological Observatory Network, in Boulder, Colorado. Dori Lynne Contreras is the curator of paleobotany at the Perot Museum of Nature and Science, in Dallas, Texas. Laura Lagomarsino is director of the herbarium and assistant professor at Louisiana State University, in Baton Rouge, Louisiana. Paula Mabee is a professor of biology at the University of South Dakota, in Vermillion, South Dakota. Robert Guralnick is an associate curator of biodiversity informatics at the Florida Museum of Natural History, at the University of Florida, in Gainesville, Florida. Marcy Revelez is a Research Associate with the Museum of Texas Tech University in Lubbock, Texas. Neil Cobb is director of the Merriam-Powell Center, at Northern Arizona University, in Flagstaff, Arizona. Katja Seltmann is director of the Cheadle Center for Biodiversity and Ecological Restoration at the University of California, Santa Barbara. Mary Catherine Aime is director of the Purdue University Herbaria and a professor at Purdue University, in West Lafayette, Indiana.
References cited
- Brewer CA, Smith D.. 2011. Vision and Change in Undergraduate Biology Education: A Call to Action. American Association for the Advancement of Science. [Google Scholar]
- Bebber DP et al.. 2010. Herbaria are a major frontier for species discovery. Proceedings of the National Academy of Sciences 107: 22169–22171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AL, Hobson KA, Branfireun BA. 2015. Rapidly increasing methyl mercury in endangered ivory gull (Pagophila eburnea) feathers over a 130 year record. Proceedings of the Royal Society B 282: 20150032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley RD, Bradley LC, Garner HJ, Baker RJ. 2014. Assessing the value of natural history collections and addressing issues regarding long-term growth and care. BioScience 64: 1150–1158. [Google Scholar]
- Carranza-Rojas J, Goeau H, Bonet P, Mata-Montero E, Joly A. 2017. Going deeper in the automated identification of herbarium specimens. BMC Evolutionary Biology 17: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook JA, Brochmann C, Talbot SL, Fedorov V, Taylor EB, Väinölä R, Hoberg E, Kholodova M, Magnusson KP, Mustonen T. 2013. Genetic perspectives on arctic biodiversity. Pages 567–591 in Meltofte H, ed. Arctic Biodiversity Assessment, Status, and Trends in Arctic Biodiveristy. Narayana Press. [Google Scholar]
- Cook J, Edwards S, Lacey EA. 2014. Natural history collections as emerging resources for innovative education. BioScience 64: 725–234. [Google Scholar]
- DuBay SG, Fuldner CC.. 2017. Bird specimens track 135 years of atmospheric black carbon and environmental policy. Proceedings of the National Academy of Science 114: 11321–11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk V. 2018. Collections-based science in the 21st century. Journal of Systematics and Evolution 56: 175–193. [Google Scholar]
- Goode GB. 1901a. The beginnings of natural history in America. Annual Report of the Board of Regents of the Smithsonian Institution, Report of the United States National Museum 1897: 357–406. [Google Scholar]
- Goode GB. 1901b. The beginnings of American Science. Annual Report of the Board of Regents of the Smithsonian Institution, Report of the United States National Museum 1897: 407–466. [Google Scholar]
- Guralnick RP et al.. 2015. Community next steps for making globally unique identifiers work for biocollections data. Zookeys 494: 133–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan L, Duhs R, Chatterjee H. 2016. Object-based learning: A powerful pedagogy for higher education. Pages 159–168 in Boys J, ed. Museums and Higher Education Working Together. Routledge. [Google Scholar]
- Hill A et al.. 2012. The notes from nature tool for unlocking biodiversity records from museum records through citizen science. Zookeys 209: 219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoberg EP, Kutz SJ, Cook JA, Galaktionov K, Haukisalmi V, Henttonen H, Laaksonen S, Makarikov A, Marcogliese DJ. 2013. Parasites. Pages529–557Meltofte H. ed. Arctic Biodiversity Assessment, Status and Trends in Arctic Biodiveristy. Narayana Press. [Google Scholar]
- Hobern D et al.. 2019. Connecting data and expertise: A new alliance for biodiversity knowledge. Biodiversity Data Journal 7: e33679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson T. 1799. A memoir on the discovery of certain bones of a quadruped of the clawed kind in the western parts of Virginia. Transactions of the American Philosophical Society 4: 246–260. [Google Scholar]
- Kao RH et al.. 2012. NEON terrestrial field observations: Designing continental‐scale, standardized sampling. Ecosphere 3: 1–17. [Google Scholar]
- Lewin HA et al.. 2018. Earth BioGenome Project: Sequencing life for the future of life. Proceedings of the National Academy of Sciences 115: 4325–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister CA, McKain MR, Li M, Bookout B, Kellogg EA. 2018Specimen-based analysis of morphology and the environment in ecologically dominant grasses: The power of the herbarium. Philosophical Transactions of the Royal Society B 374: 20170403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel M. 1926. A Bibliography of American Natural History, vol. 2: The Pioneer Century, 1769–1865. The Premier Publishing Company. [Google Scholar]
- Melber LM, Abraham LM.. 2002. Science education in U.S. natural history museums: A historical perspective. Science and Education 11: 45–54. [Google Scholar]
- Monfils A, Powers KE, Marshall CJ, Martine CT, Smith JF, Prather LA. 2017. Natural history collections: Teaching about biodiversity across time, space, and digital platforms. Southeastern Naturalist 16: 47–57. [Google Scholar]
- Page LM, MacFadden BJ, Forte JA, Soltis PS, Riccardi G. 2015. Digitization of biodiversity collections reveals biggest data on biodiversity. BioScience 65: 841–842. [Google Scholar]
- Palkovacs EP, Kinnison MT, Correa C, Dalton CM, Hendry AP. 2012. Fates beyond traits: Ecological consequences of human‐induced trait change. Evolutionary Applications 5: 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrelli D, Ciolfi L, van Dijk D, Hornecker E, Not E, Schmidt A. 2013. Integrating material and digital: A new way for cultural heritage. Interactions 20: 58–63. [Google Scholar]
- [NRC] National Research Council 2013. Next Generation Science Standards: For States, by States. National Academies Press. [PubMed] [Google Scholar]
- Nelson G, Ellis S.. 2018. The history and impact of digitization and digital data mobilization on biodiversity research. Philosophical Transactions of the Royal Society B 374: 20170391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samy AM, Elaagip AH, Kenawy MA, Ayres CFJ, Peterson AT, Soliman DE. 2016. Climate change influences on the global potential distribution of the mosquito Culex quinquefasciatus, vector of west nile virus and lymphatic filariasis. PLOS ONE 11 (art. e0163863). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis PS. 2017. Digitization of herbaria enables novel research. American Journal of Botany 104: 1281–1284. [DOI] [PubMed] [Google Scholar]
- Schindel DE, Cook JA.. 2018. The next generation of natural history collections. PLOS Biology 16 (art. e2006125). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trejo-Salazar R-E, Eguiarte LE, Suro-Piñera D. 2016. Save our bats, save our tequila: Industry and science join forces to help bats and agaves. Natural Areas Journal 36: 523–530. [Google Scholar]
- Tewksbury JJ, Anderson JGT, Bakker JD. 2014. Natural history's place in science and society. BioScience 64: 300–310. [Google Scholar]
- Thiers B. 2019. Index Herbariorum. http://sweetgum.nybg.org/science/ih. [Google Scholar]
- Webster MS, ed. 2017. The Extended Specimen: Emerging Frontiers in Collections-Based Ornithological Research. CRC Press. [Google Scholar]