Table 4. Structure refinement.
Values in parentheses are for the outer shell.
| GR839 | GSK513 | GW197 | |
|---|---|---|---|
| Fragment structure |
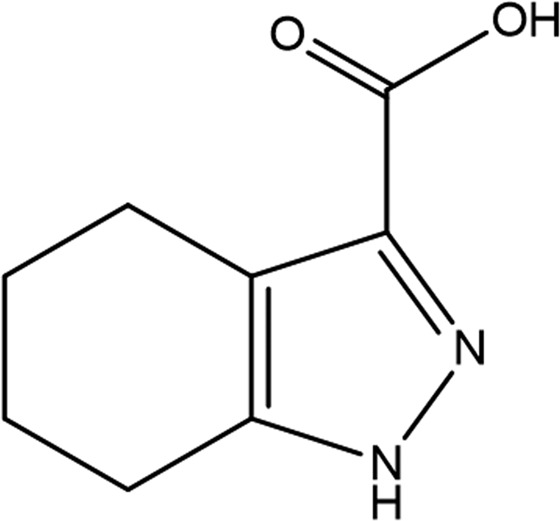
|
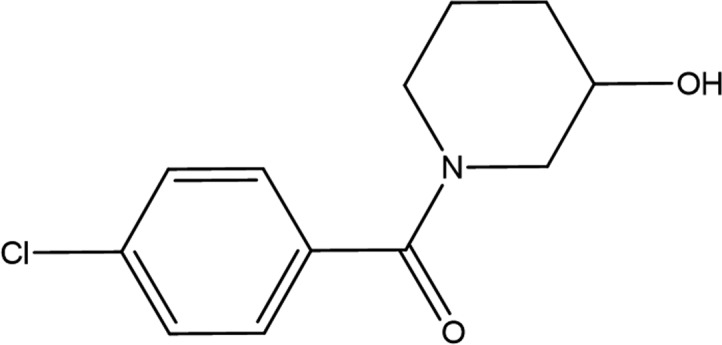
|
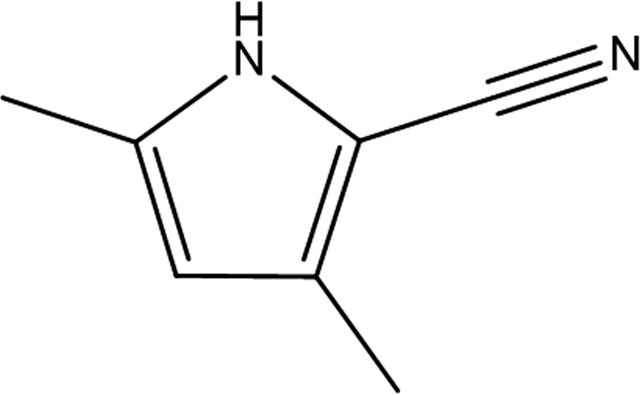
|
| Resolution range (Å) | 20.09–1.84 (1.91–1.84) | 23.87–1.65 (1.67–1.65) | |
| Completeness (%) | 80.5 (14.1) | 79.5 (30.7) | |
| No. of reflections, working set | 35922 (757) | 49773 (1048) | |
| No. of reflections, test set | 1911 (32) | 2625 (37) | |
| Final R cryst | 0.171 (0.2097) | 0.183 (0.2197) | |
| Final R free | 0.204 (0.2914) | 0.212 (0.2420) | |
| Cruickshank DPI | 0.143 | 0.108 | |
| No. of non-H atoms | |||
| Protein | 3898 | 3870 | |
| Ion | 1 | 1 | |
| Ligand | 28 | 9 | |
| Water | 368 | 324 | |
| Total | 4295 | 4204 | |
| R.m.s. deviations | |||
| Bonds (Å) | 0.01 | 0.01 | |
| Angles (°) | 1.0 | 1.6 | |
| Average B factors (Å2) | |||
| Protein | 23 | 31 | |
| Ion | 23 | 26 | |
| Ligand | 51 | 30 | |
| Water | 32 | 39 | |
| Ramachandran plot† | |||
| Favoured regions (%) | 98.60 | 99.08 | |
| Additionally allowed (%) | 1.40 | 0.69 | |
| Outliers (%) | 0 | 0.23 | |
| PDB code | 6szg | 6szh | |
Values determined by MolProbity (Chen et al., 2010 ▸).
