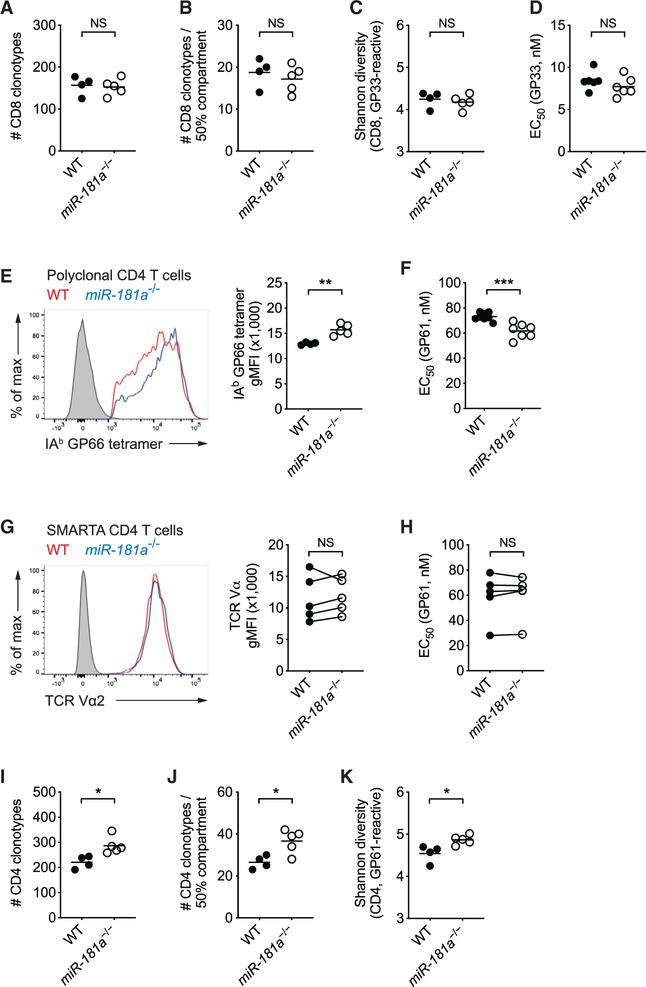
Figure 3. miR-181a Deficiency Selects a More Diverse and Higher-Affinity Virus-Specific CD4 T Cell Repertoire.
WT and miR-181a−/− mice were infected with LCMV. (A–C) TRB genes from splenic Db GP33 tetramer+ YFP+ CD8 T cells on day 8 were sequenced. Total number of unique TRB sequences (A), the number of top frequent clones making up 50% of the entire repertoire (B), and the Shannon diversity index (C) are shown.
(D) Splenocytes were restimulated with a range of GP33 peptide concentrations. Dot plot shows the effective GP33 peptide concentration required to elicit a half-maximal IFNγ production.
(E) Representative histogram of tetramer binding intensity of IAb GP66 tetramer+ YFP+ CD4 T cells (left) and summary data of tetramer mean fluorescence intensity (MFI) from one experiment (right). Filled gray in histogram indicates naive CD4 T cells.
(F) Dot plot shows the effective GP61 peptide concentration required to elicit a half-maximal IFNγ production after in vitro restimulation.
(G and H) WT and miR-181a-deficient SMARTA cells were co-transferred into WT mice that were then infected with LCMV. MFI expression of the SMARTA TCR as histogram (G, left), summary plot from 5 mice in each group (G, right), and half-maximal effective concentration (EC50) of IFNγ production by SMARTA cells after peptide restimulation in vitro (H) on day 8. (I–K) TRB sequences from splenic IAb GP66 tetramer+ YFP+ CD4 T cells were determined on day 8. Total number of unique TRB sequences (I), the number of top frequent clones making up 50% of the entire repertoire (J), and the Shannon diversity index (K) are shown.
Data are from one experiment with 4–5 mice per group (A–C), representative of two independent experiments with 4–5 mice per group (D, E, and G–K) or pooled from two independent experiments with 7–8 mice per group(F). Unless stated otherwise, data are presented as means. Statistical significance by two-tailed unpaired (A–F and I–K) or paired (G and H) t test. *p < 0.05; **p <0.01; ***p < 0.001; NS, not significant.