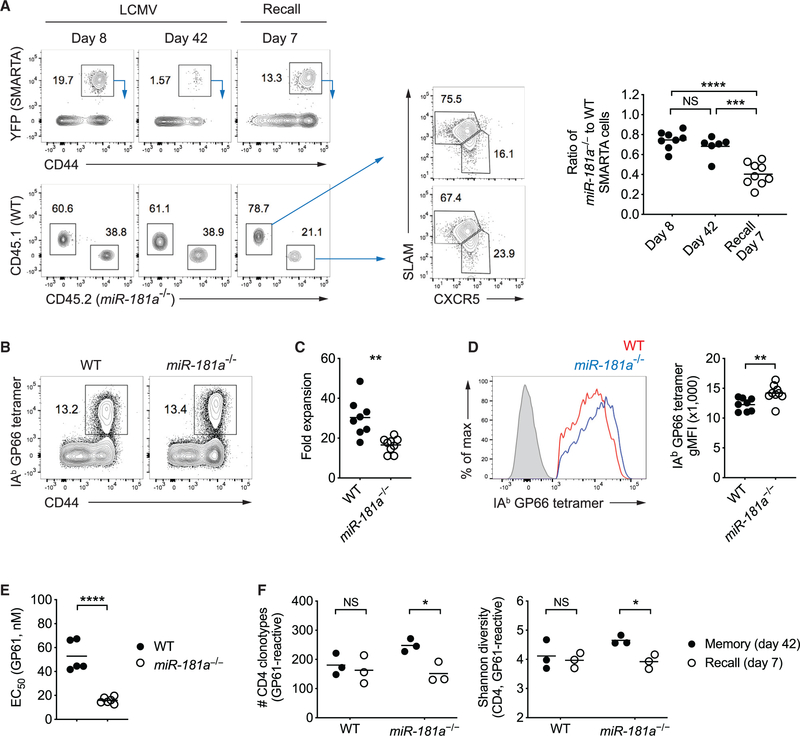
Figure 7. miR-181a Is Required for Secondary Expansion of Memory CD4 T Cells.
(A) Equal numbers of congenically marked WT and miR-181a−/−YFP+ SMARTA CD4 T cells were co-transferred into B6 mice infected with LCMV 1 day later. On day 50 after infection, immune mice were rechallenged with Lm-gp61. Representative flow plots show YFP+ SMARTA cells gated on CD4 T cells (top left) and the relative frequencies of WT and miR-181a−/− SMARTA cells (bottom left) in the spleen at indicated time points after infection and SLAM and CXCR5 expression on WT and miR-181a−/− SMARTA CD4 T cells on day 7 after recall (middle). Dot plots on the right show the ratios of miR-181a−/− to WT SMARTA CD4 T cells.
(B–F) WT and miR-181a−/− mice were infected with LCMV. On day 80 after infection, immune mice were rechallenged with Lm-gp61 and cells were harvested on day 7 at the peak of the recall response. (B) Representative flow plots of IAb GP66 tetramer+ cells gated on YFP+ CD4 T cells. (C) Fold expansion of IAb GP66 tetramer+ memory CD4 T cells on day 7 after rechallenge compared to before infection. (D) Representative histogram of tetramer binding intensity of IAb GP66 tetramer+ YFP+ CD4 T cells (left) and dot plot of tetramer MFI (right). Filled gray in the histogram indicates naive CD4 T cells. (E) Effective GP61 peptide concentration required to elicit a half-maximal IFNγ production. (F) TRB genes from IAb GP66 tetramer+ YFP+ CD4 T cells in the spleen were sequenced. Dot plots show the total number of unique TRB sequences (left) and the Shannon diversity index (right).
Data are pooled from two independent experiments with 5–9 mice per group (A–E) or are from one experiment with 3 mice per group (F). Unless stated otherwise, data are presented as means. Statistical significance by two-tailed unpaired t test. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; NS, not significant.