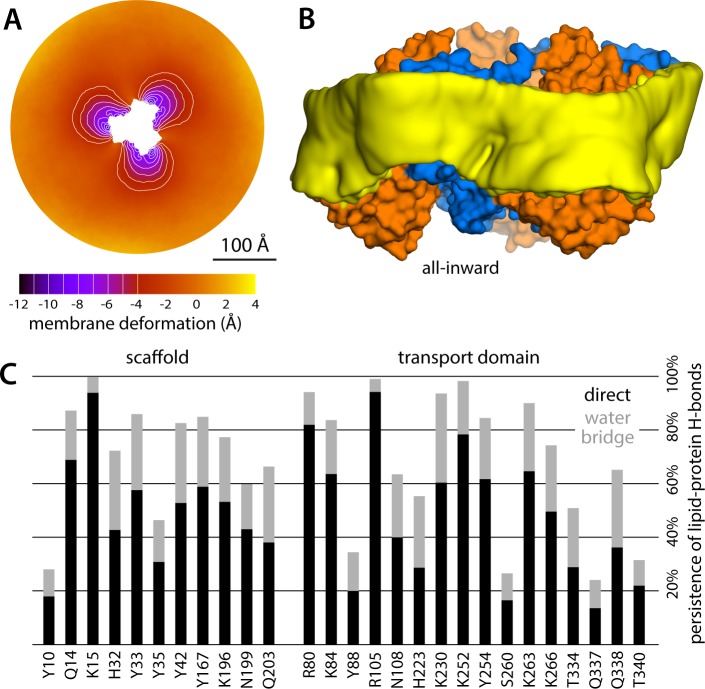
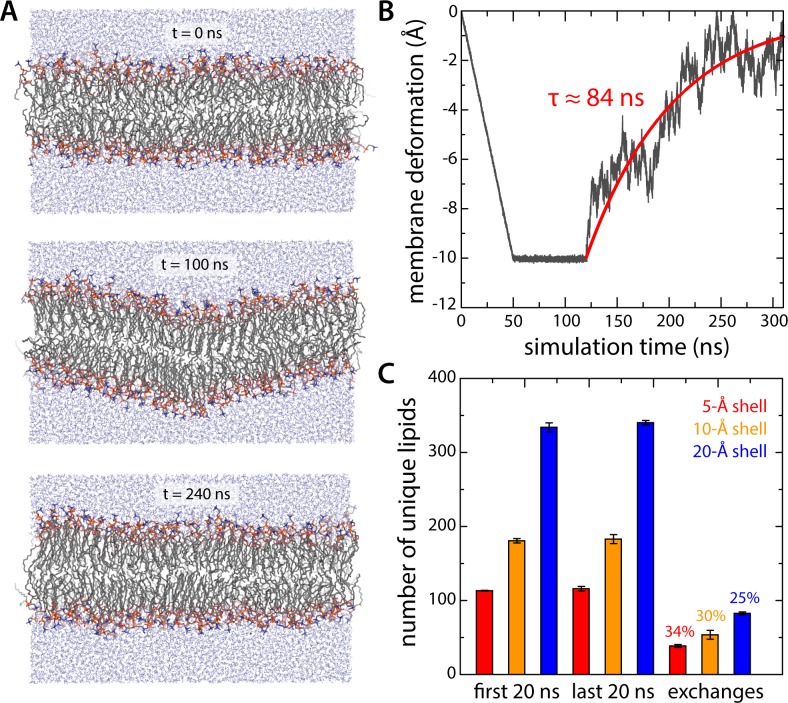
(
A, B) All-atom simulation of the relaxation of an artificial 10 Å deflection in the membrane mid-plane. Given the large system size of the all-atom simulation system for all-inward Glt
Ph (~3,000,000 atoms), we sought to determine the minimum sampling time required to evaluate whether the membrane deformation induced by the protein in coarse-grained simulations was compatible with this more accurate representation. To this end we constructed a simulation system consisting of a 640-DPPC lipid bilayer (323 K) and ~32,700 TIP3P water molecules, enclosed in a periodic box of ~140 × 140×80 Å. After equilibration, an artificial deflection of the membrane mid-plane was created in this bilayer whose magnitude is comparable to that observed in the simulations of inward-facing Glt
Ph (see
Figure 2A). To create this perturbation, lipid molecules inside a cylinder of ~10 Å in radius were progressively displaced perpendicularly to the plane of the membrane by applying a harmonic restraint with a moving center and force constant of 100 kcal/mol Å
−2. The rate of movement of the restraint center was such that after 50 ns of simulation the vertical deflection was 10 Å; this value was then sustained for 70 ns, using the same restraint, now stationary. An unrestrained simulation of ~200 ns was carried out thereafter to evaluate the persistence or relaxation of the membrane deformation, during which we observed that the membrane relaxed fully within ~150 ns. Accordingly, each of the three all-atom trajectories calculated for inward-facing Glt
Ph was 150 ns long. (
C) Lipid dynamics in the all-atom simulations of inward-facing Glt
Ph. The plot quantifies the number of lipid molecules that reside in three different shells around the protein (of 5, 10 or 20 Å width – red, orange and blue, respectively) for at least 50% of a 20-ns window at the beginning of the simulations (
N = 3). The same values are given for a 20-ns window at the end of the simulations. The number of lipid molecules found in the latter set but not the former gives the number of exchanges between lipid molecules inside the shell and those further away from the protein.