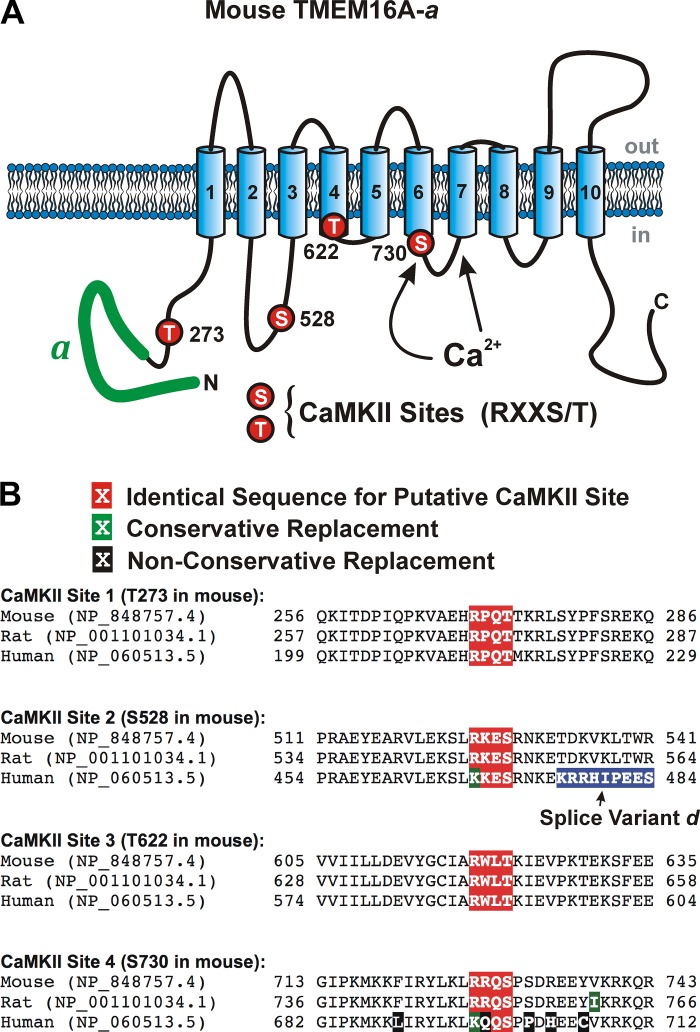
Fig. 7.
Proposed membrane topology and location of the four putative calmodulin-dependent protein kinase II (CaMKII) phosphorylation sites identified on mouse TMEM16A. A: proposed secondary structure of mouse TMEM16A highlighting the alternatively spliced variant “a” located in the NH2 terminus (thick green solid line) and approximate location of the proposed binding sites for Ca2+ based on the structure proposed by Paulino et al. (30), and the position of the four consensus sites for phosphorylation by CaMKII that obey the basic sequence RXXS/T (T273, S528, T561, and S730 all labeled with red circles). B: sequence homology among mouse, rat, and human TMEM16A in the region surrounding each of the four identified potential CaMKII phosphorylation targets. The putative CaMKII RXXT/S sequence and conservative and nonconservative amino acid replacements are highlighted in white font over a red, green, or black background, respectively, as shown above the sequences. The amino acids highlighted in blue for the human sequence corresponds to splice variant “d,” which is not present in the mouse and rat sequences defined by the two National Center for Biotechnology Information protein accession numbers. However, the human sequence downstream of splice variant “d” (not shown) defined by accession no. NP_060513.5 is identical to the remaining sequence shown for mouse and rat (TDKVKLTWR).