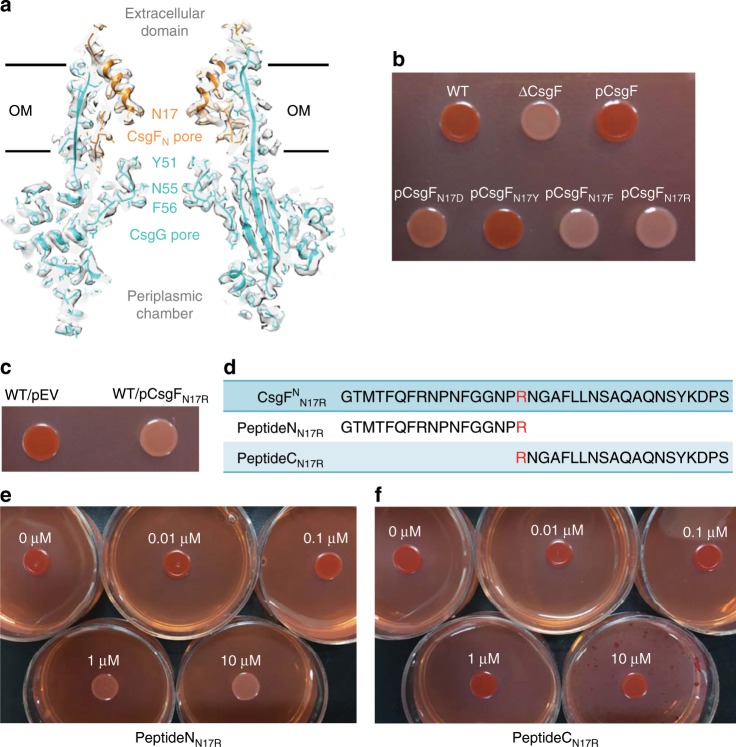
Fig. 2. Dual-pore structure of CsgFG complex and analysis of peptide inhibition.
a Section view of CsgFG complex. The atomic model was superimposed with the corresponding cryoEM density map. Models of CsgG and CsgF are in cyan and orange, respectively. b Congo red secretion assay for mutations of the asparagine residue of CsgFN into four different kinds of amino acids in the ∆CsgF strain. Source data are provided as a Source Data file. c Congo red secretion assay for the N17R-mutated CsgF in the wild type (WT) strain. Source data are provided as a Source Data file. d Schematic diagram of the synthesized peptides, PeptideNN17R and PeptideCN17R. The N17R mutation is shown in red. e Congo red secretion assay for the WT strain treated with PeptideNN17R at concentrations ranging from 0.01 to 10 µM. Source data are provided as a Source Data file. f Congo red secretion assay for the WT strain treated with PeptideCN17R at concentrations ranging from 0.01 to 10 µM. Source data are provided as a Source Data file.