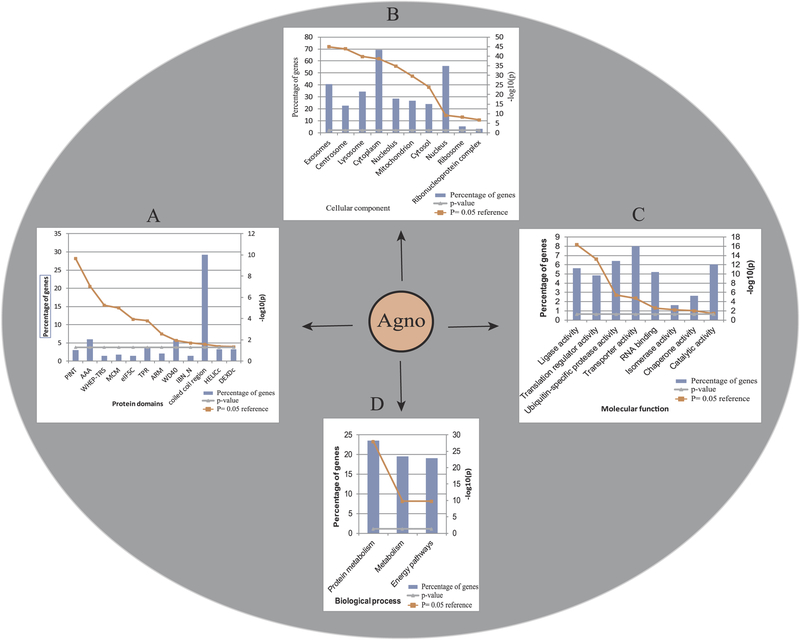
Fig. 4. Enrichment and classification of Agno-targeted host proteins using FunRich software.
Agno-targeted host proteins were further analyzed by using “FunRich”, based on the p = 0.05 reference value. Agno-interacting proteins were grouped together on the bases of having specific (A) “protein domains”, (B) as those integrated into various “cellular components” (exosomes, centrosomes, lysosomes, cytoplasm, nucleolus, mitochondrion, cytosol, nucleus, ribosome and nucleoprotein complexes), (C) those associated with various “molecular functions” (ligase activity, translation regulator activity, ubiquitin specific protease activity, transporter activity, RNA binding, isomerase activity, chaperone activity and catalytic activity) and (D) those involved in various “biological processes” (protein metabolism, metabolism and energy pathways). Percentage of genes is defined as the ratio of the host proteins that interact with Agno to the available number of proteins present in the FunRich data in that specific category.