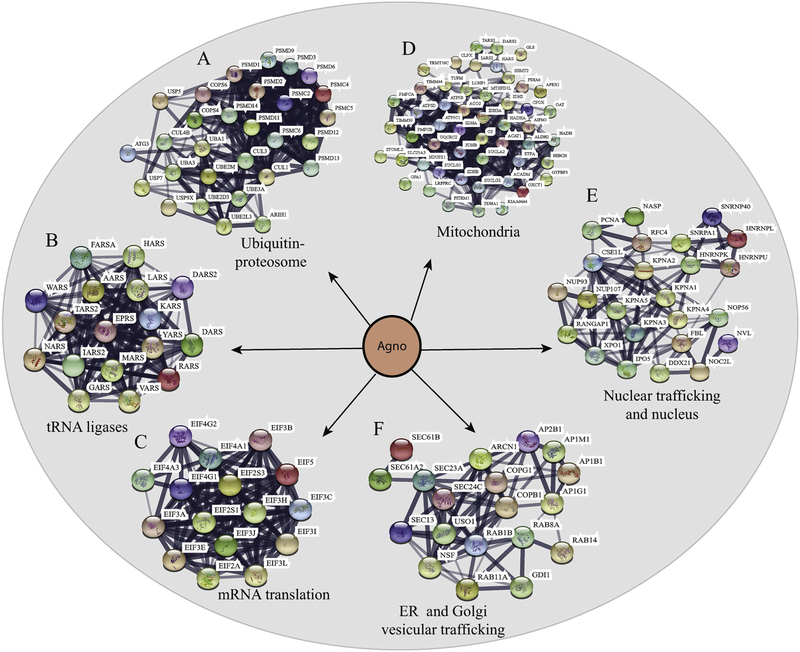
Fig. 5. Interactome maps for the selected host-cell protein networks targeted by Agno using “STRING database”.
The host-cell protein networks involved in (A) protein degradation pathways (ubiquitinproteasome complexes), (B) synthesis of tRNA (tRNA synthases) and (C) mRNA translation pathways. (DEF) Establishment of the interactome maps for the organelle-integrated proteins targeted by Agno using lism, m databases), (B)mitochondria-integrated proteins, (E) nuclear proteins and the proteins involved in nuclear trafficking networks, (F) endoplasmic reticulum (ER)- and Golgi network vesicular trafficking.