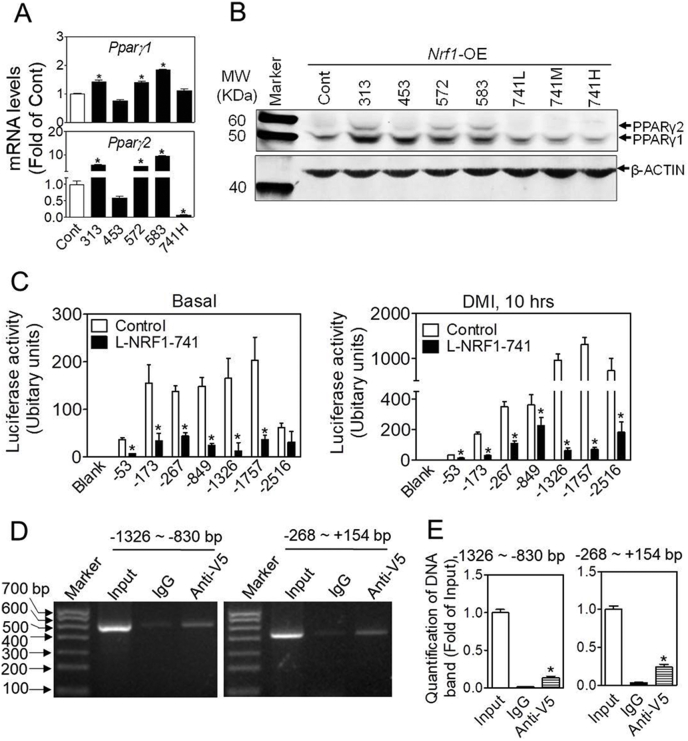
Fig. 6.
Overexpression of L-NRF1-741 suppresses the expression of PPARγ in 3T3-L1 cells. (A) The mRNA expression of Pparγ in control and various Nrf1-OE cells under normal culture condition. Cont (Control), 313, 453, 572, 583, and 741 refer to the cells overexpressing indicated isoforms of NRF1. *p < 0.05 vs. Control. n = 3-5. (B) Representative image of Western blot detecting PPARγ1/2 in control and various Nrf1-OE cells under basal condition. 741L/M/H refer to the cells overexpressing L-NRF1-741 using low (L), medium (M), and high (H) concentrations of lentiviral vector. n = 3. (C) The activity of Pparγ2 promoter-reporter assay. Control and NRF1-741 cells were transfected with the Pparγ2 promoter-luciferase constructs. After 2 days, the transfected cells were treated with basal growth medium (left panel) and DMI differentiation media (right panel) for 10 h. Blank, no insert of reporter; −53, −173, −267, −849, −1326, −1757 and −2516 represent the reporters driven by the 5′-truncated Pparγ2 promoter. n = 3–6; *p < 0.05 vs. Control expressing the same reporter. (D) Representative images of ChIP analysis for the interaction between NRF1-741 and promoter region of Pparg2 gene. 3T3-L1 cells overexpressing V5-tagged NRF1-741 were treated with DMI and 10 μM MG132 for 6 h when reaching 90% confluence, followed by ChIP assay. Input, enriched DNA by mouse IgG and V5 antibody were amplified by PCR with 8 pairss of primers designed to flank the continuous 3,000 bp range in the upstream of Pparγ2 gene. Non-immunoprecipitated chromatin (2%) was used as input control. PCR products were resolved with 1% DNA gel. n = 3. (E) Quantification of DNA bands in Fig. D. n = 3; *p < 0.05 vs. 2% input. (E) Quantification of DNA bands in Fig. D. n = 3; *p < 0.05 vs. 2% input.