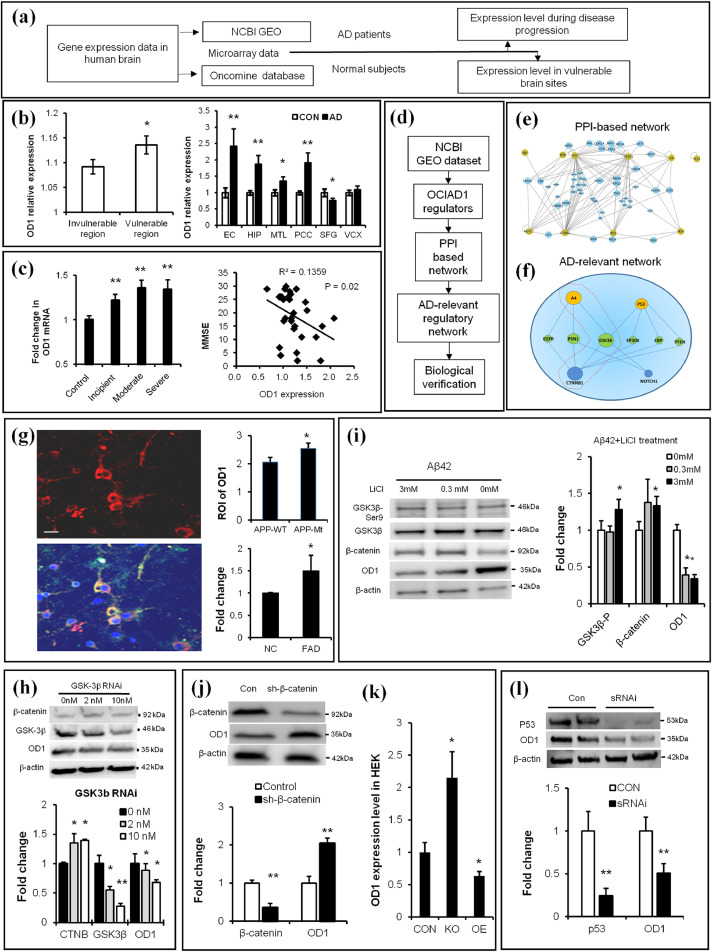
Fig. 2.
Brain OCIAD1 level is associated with AD development and under regulation of Aβ/GSK3β/p53 signals. (a) Workflow for analysis of OD1(OCIAD1) gene expression in human brain. (b-c) Higher expression level of OD1 was shown in the vulnerable brain sites in normal human subjects (left, in b) and AD patients (right, in b) while the increased level of OD1 correlates with severity of AD (c). (d-e) OD1 regulators were derived from GEO database to build PPI-based connection network (e) and generate AD-relevant regulatory network (f). (See also Table 1). (g-h) Elevated APP (g) or knockdown GSK-3β (h) change OD1 level in cells. APP-staining (red), OD1-staining (green), DAPI-staining (blue). APP mutant (APP-Mt), APP wild type (APP-Wt), wild type (NC), transgenic mice (FAD), knockdown GSK3β via sRNAi (GSK3β RNAi). (See also Fig. S2a-2c). (i) Chemical intervention of Aβ-GSK-3β signal affected levels of β-catenin and OD1 in cultured primary neurons. (j) Modulating β-catenin changed the OD1 level in HEK293 cells as shown in GEO data. (k) Knockdown (KO), over-expression (OE), knockdown β-catenin via shRNAi (sh-β-catenin). (l) Knockdown p53 reduces OD1 level in HEK293. Knockdown p53 via sRNAi (sRNAi). (See also Fig. S2d-2e). Data are presented as mean±SEM in (b–c, g–l). *P < 0.05, **P < 0.01 vs. control in (b) (student t-test, left panel, n = 32; right panel, n = 5), in c (one-way ANOVA, left panel, n = 3; Pearson Correlation Coefficient, right panel, n = 30), in h-l (student t-test, n = 3 in h-j, l, n = 6 in k),.*P < 0.05 vs. wild type in g (student t-test, n = 33 in APPWT, n = 45 in APPMt, and n = 3 in NC and FAD). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)