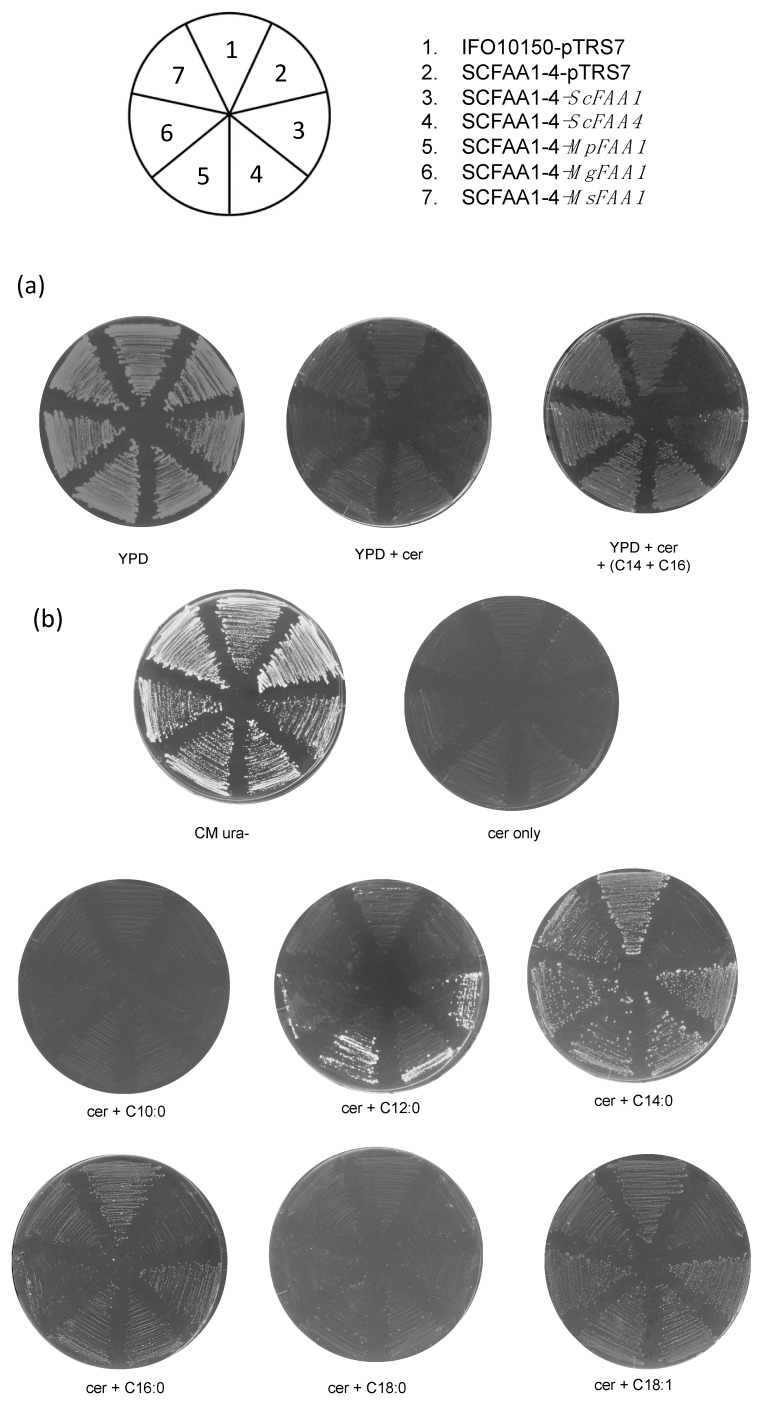
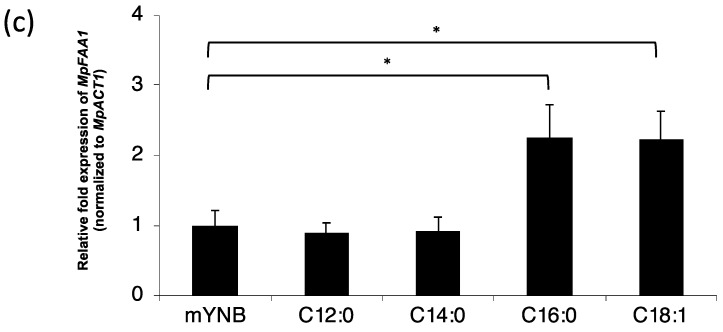
Figure 2.
(a) Complementation analysis and substrate specificity determination for MpFAA1, MgFAA1, and MsFAA1. S. cerevisiae IFO10150-pTRS7 (FAA1FAA4) and its FAA1 and FAA4 deletion mutant (SCFAA1-4--pTRS7) as well as SCFAA1-4-ScFAA1, SCFAA1-4-ScFAA4, SCFAA1-4-MpFAA1, SCFAA1-4-MgFAA1, and SCFAA1-4-MsFAA1 were streaked on YPD agar plates containing cerulenin and C14:0 and C16:0 fatty acids. (b) Growth on CM ura− agar plates containing C10:0, C12:0, C14:0, C16:0, C18:0, and C18:1 (0.5 mM) and supplemented with 5 μg/mL cerulenin and cultured at 30 °C for 5–10 days. CM ura− agar plates containing cerulenin without fatty acids were used as controls. (c) Levels of MpFAA1 transcripts in mYNB medium containing various fatty acids. Malassezia pachydermatis 1879NT was grown in mDixon liquid medium until early/mid-log phase, transferred to mYNB medium containing 5 mM fatty acid (C10:0, C12:0, C14:0, C16:0, C18:0, and C18:1), and incubated for 6 h at 32 °C. RNAs were extracted, reverse transcribed to cDNA, and then analyzed to determine gene expression level by qRT-PCR relative to the expression of MpACT1 (n = 3 experiments, each done in duplicate; * p < 0.05).