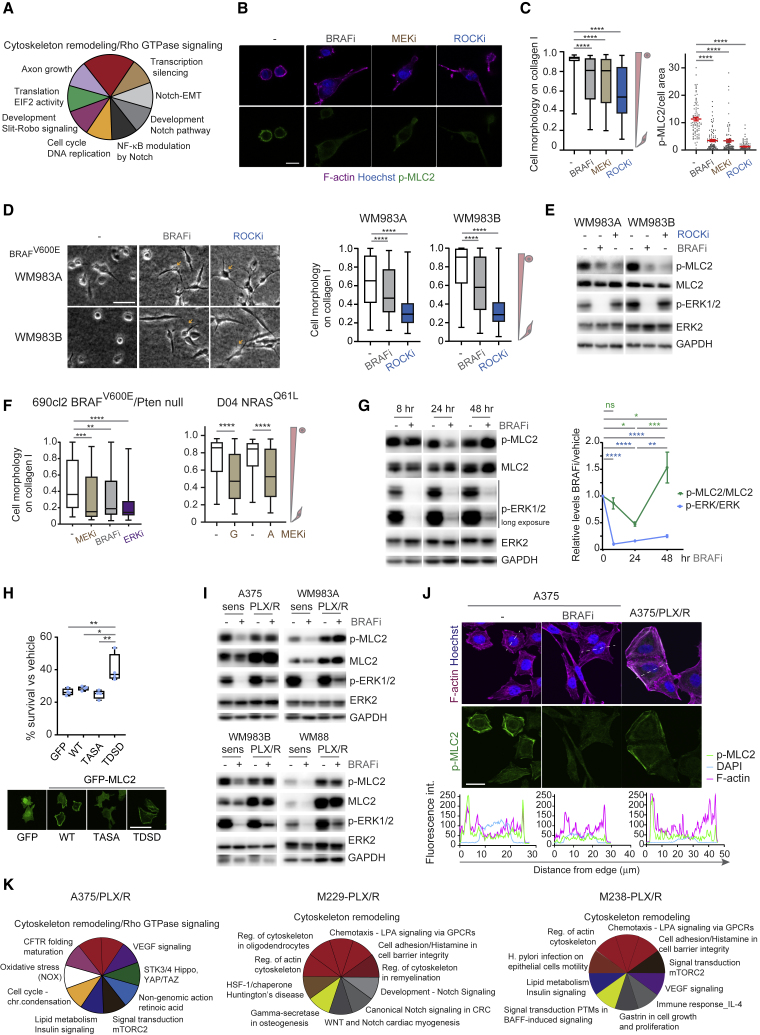
Figure 1.
MAPK Regulates Myosin II Activity in Melanoma
(A) The 10 most enriched pathways in A375 cells after MEKi treatment compared with untreated cells from phospho-proteome data.
(B) p-MLC2 and F-actin confocal images of A375M2 cells on collagen I after treatment (BRAFi PLX4720, MEKi trametinib, ROCKi GSK269962A). Scale bar, 25 μm.
(C) Quantification of cell morphology and p-MLC2 by immunofluorescence from (B). Left, boxplot (n > 200 cells pooled from 3 experiments); right, mean ± SEM (n = 90 cells [dots] pooled from 3 experiments).
(D) Images and quantification of cell morphology on collagen I after treatment (BRAFi PLX4720, ROCKi GSK269962A) (n > 346 cells pooled from 2 experiments). Arrows show collapsed phenotype. Scale bar, 100 μm.
(E) p-MLC2 and p-ERK1/2 immunoblots from (D).
(F) Cell morphology on collagen I after treatment (690cl2, MEKi PD184352, BRAFi PLX4032, ERKi SCH772984, n = 50 cells; D04, MEKi GSK1120212, AZD6244, n = 125–150 cells).
(G) p-MLC2 and p-ERK1/2 levels after PLX4720 treatment (n = 5, mean ± SEM).
(H) Survival of A375 cells stably overexpressing wild type (WT), constitutively inactive TASA, or constitutively active TDSD MLC2 a after 5-day treatment with 0.1 μM PLX4720 (n = 4). Confocal images of GFP-MLC2. Scale bar, 50 μm.
(I) p-MLC2 and p-ERK1/2 immunoblots after PLX4720 treatment.
(J) p-MLC2 and F-actin confocal images (BRAFi PLX4720). Scale bar, 25 μm. Representative fluorescence intensity line scans (dashed lines in image) below.
(K) The 10 most enriched pathways in BRAFi-resistant A375/PLX/R (Girotti et al., 2013), M229-PLX/R, and M238-PLX/R cells (Titz et al., 2016) compared with parental cell lines from phospho-proteome data.
(A–F, I, and J) 24 h treatment.
(C, D, F, and H) Boxplots show median (center line); interquartile range (box); min-max (whiskers). p values by Kruskal-Wallis with Dunn's correction (C, D, and F), one-way ANOVA with Tukey's (H) or Benjamini, Krieger, and Yekutieli correction (G), ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001. See also Figure S1 and Tables S1 and S2.