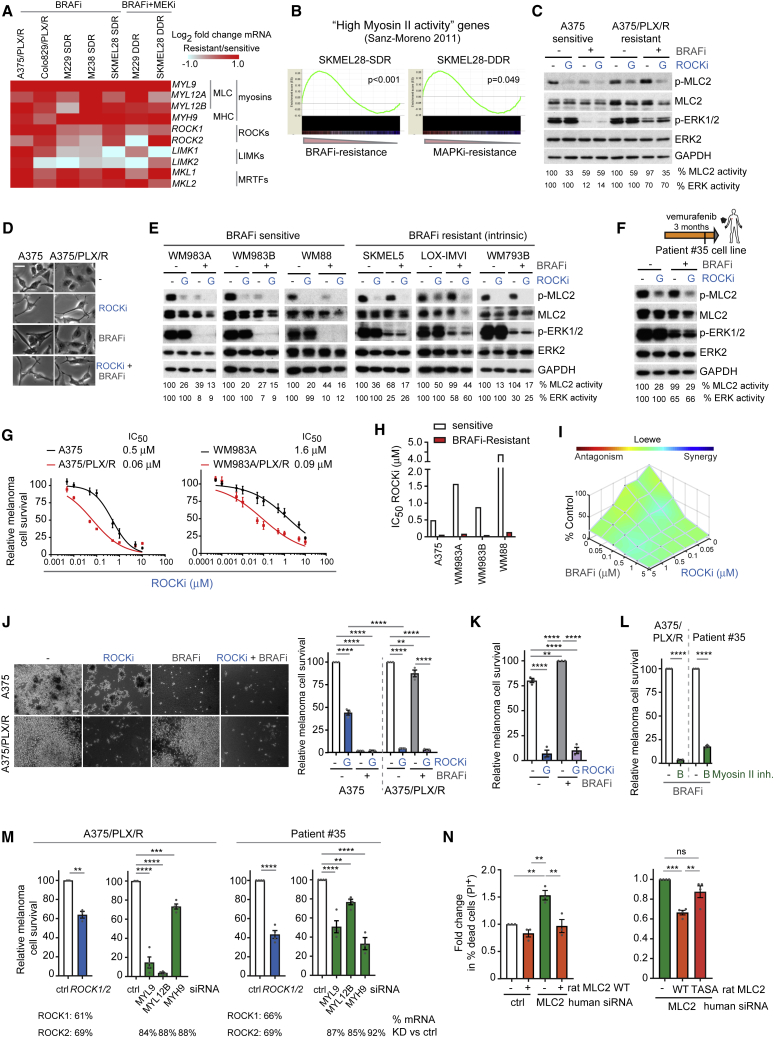
Figure 3.
Survival of Targeted Therapy-Resistant Melanomas Is Dependent on ROCK-Driven Myosin II Activity
(A) Fold change in mRNA levels of ROCK-myosin II pathway genes in A375/PLX/R, Colo829/PLX/R by qRT-PCR (n = 3); and from published RNA sequencing data (Song et al., 2017).
(B) GSEA comparing high myosin II activity signature (Sanz-Moreno et al., 2011) to resistant cell lines (Song et al., 2017). Nominal p values shown, false discovery rate (FDR) < 0.2.
(C) p-MLC2 and p-ERK1/2 immunoblots after 24 h treatment.
(D) Images of cells from (C). Scale bar, 50 μm.
(E and F) p-MLC2 and p-ERK1/2 immunoblots of sensitive and intrinsically resistant cells (E); and patient no. 35 cells (F) after 24 h treatment (8 h for WM88). Vertical line in diagram (F): cell line establishment.
(G) Survival and half maximal inhibitory concentration (IC50) values after a 3-day treatment (n = 3).
(H) IC50 values for GSK269962A.
(I) Cell survival as synergy graph of A375 cells treated for 3 days (n = 4).
(J) Images and quantification of cell survival on collagen I for 9 days (n = 3). Scale bar, 100 μm.
(K) Survival of patient no. 35 cells after 10 days (n = 3).
(L) Survival after a 5- to 10-day blebbistatin and PLX4720 treatment (n = 3).
(M) Survival 8 days after gene depletion by RNAi (n = 3; n = 4 A375/PLX/R myosin genes, patient no. 35 MYL12B, ROCK1/2). mRNA KD (percentage decrease versus control) by RT-PCR shown.
(N) Cell death in A375/PLX/R cells 3 days after transient MLC2 KD and rescue with rat MLC2 WT or TASA (n = 3, left graph; n = 4, right graph).
(C–K) ROCKi GSK269962A, BRAFi PLX4720.
Graphs show mean ± SEM and individual data points (circle). p values by one-way ANOVA with Tukey's (J, K, and N) or Dunnett's correction (M, myosin genes); t test with Welch's correction (L and M, ROCK), ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; n.s., not significant. See also Figures S2 and S3.