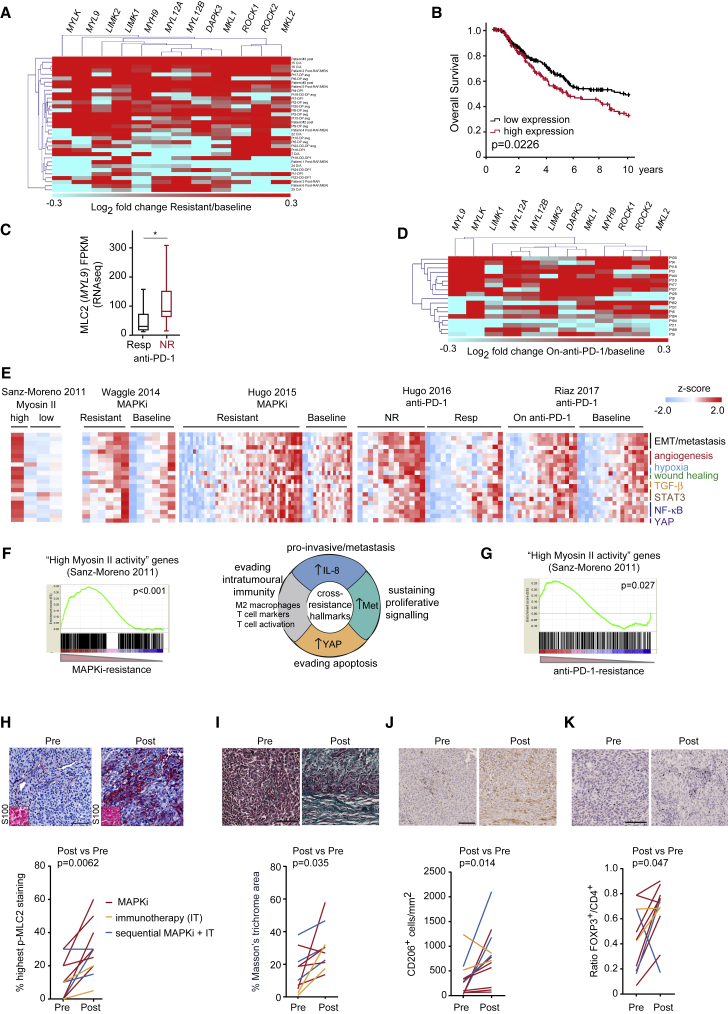
Figure 4.
High Myosin II Levels Identify Therapy-Resistant Melanomas in Human Samples
(A) Heatmap of fold change in expression of ROCK-myosin II pathway genes in MAPKi-resistant versus baseline patient samples from (Hugo et al., 2015, Kwong et al., 2015, Sun et al., 2014, Wagle et al., 2014).
(B) Kaplan-Meier overall survival from The Cancer Genome Atlas according to expression of ROCK-myosin II genes (listed in A) (n = 389 melanoma patients).
(C) MYL9 mRNA in Resp (n = 15) and NR (n = 13) anti-PD-1 patients from (Hugo et al., 2016). Boxplot: median (center line); interquartile range (box); min-max (whiskers).
(D) Heatmap of fold change in expression of ROCK-myosin II genes in on-anti-PD-1 versus baseline patient samples (Riaz et al., 2017).
(E) Heatmaps show ssGSEA of cross-resistance gene signatures (NR, non-responder; Resp, responder).
(F and G) GSEA comparing “high myosin II activity” signature (Sanz-Moreno et al., 2011) to a subset of MAPKi-resistant patient samples from (Hugo et al., 2015) (F) or anti-PD-1/NR samples (Hugo et al., 2016) (G). Chart pie in (F) with cross-resistance hallmarks from (Hugo et al., 2015). Nominal p values shown, FDR < 0.001 (F) and 0.145 (G).
(H–K) Images (patient no. 17) and quantification in 12 paired samples before and after therapies (including those in Figures S4E and S4F) of: p-MLC2 (% cells with highest score), melanoma marker S100 (inset) (H); Masson's trichrome staining (percentage stained area/section) (I); CD206+ cells (J); FOXP3+ cells (K). Scale bars, 100 μm.
p values by Mann-Whitney test (C, H–K). See also Figure S4 and Tables S4, S5, and S6.