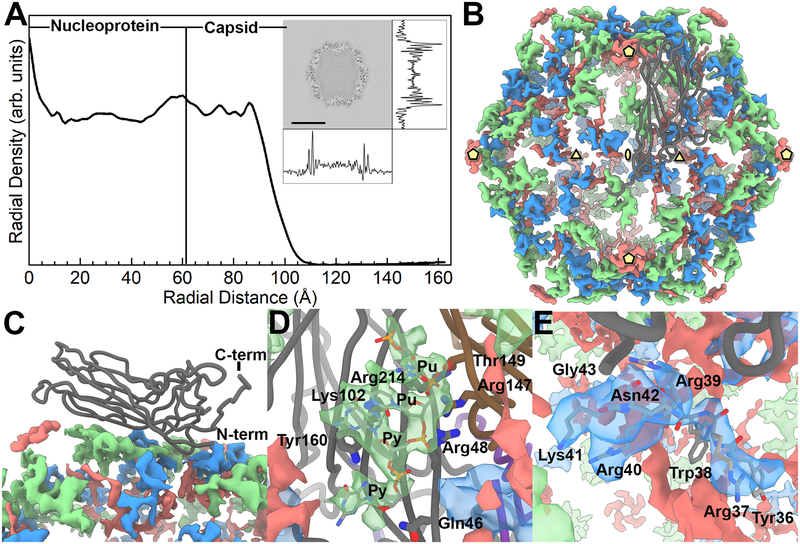
Figure 4. The inner content of the PCV2 capsid.
A) A radial profile of the PCV2d cryo-EM molecular envelope. The capsid interior and shell are identified by assessing the cryo-EM molecular envelope. The inset is a central slice extracted from the cryo-EM molecular envelope, with the density trace of pixel values calculated in the horizontal and vertical directions. The radial profile demonstrates that number of voxels within the capsid is comparable to the capsid shell; thus, a substantial amount of material is located within the capsid interior. B) Strong difference peaks identified in the inner capsid. The icosahedral 5-, 3- and 2-fold axes of symmetry are identified by yellow pentagons, triangles and ellipses, respectively. A CP subunit is shown as a dark grey tube. We interpret the green colored difference peak to be a Pu-Pu-Py-Py tetranucleotide, the blue colored difference peak to be amino acids 36–41 of the PCV2 N-terminus, and the red colored difference peak to be “unidentified”. C) Side view showing the CP subunit and the difference peaks. D) Close up of the tetranucleotide that has been modeled into the difference peak (green) located near the 3-fold axes of symmetry, and the CP amino acids in proximity. Gln46 (strand B), Arg48 (strand B), Lys102 (strand D), and Arg214 (strand I) of one subunit, and Thr149 (strand F) and Arg147 (strand F) from a neighboring subunit form hydrogen bonds and electrostatic interaction with the phosphate backbones of the tetranucleotide. Tyr160 (strand I) forms π-bond overlap with the first Py in the tetranucleotide. E) Close up of the PCV2 N-terminus modeled into the difference peak (blue) located near the 3-fold axes of symmetry. Amino acids 36–42 are labeled.