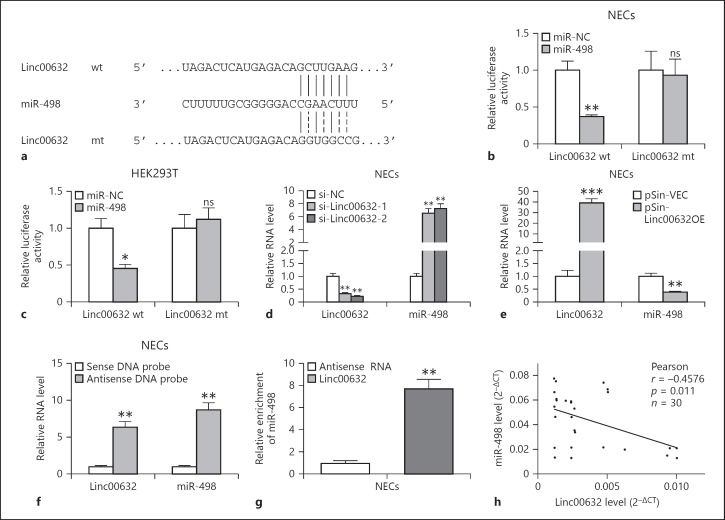
Fig. 3.
Linc00632 negatively regulated miR-498 as an RNA sponge. a The prediction for miR-498-binding sites on Linc00632 transcript and schematic of luciferase reporter vector constructs Linc00632 wt and the miR-498-binding site Linc00632 mt one. b, c The luciferase activities in NECs and HEK293T cells co-transfected with miR-498 or miR-NC mimics and luciferase reporters containing Linc00632 wt or Linc00632 mt. Data are presented as the relative ratio of hRluc luciferase activity to hluc+ luciferase activity. d, e The expression levels of Linc00632 and miR-498 in NECs transfected with Linc00632 siRNAs (si-Linc00632–1, si-Linc00632–2) or negative control (si-NC) and Linc00632 overexpression plasmid (pSin-Linc00632OE) or empty vector (pSin-VEC) were measured by qRT-PCR. f Lysates from NECs were incubated with in vitro-synthesized biotin-labeled sense or antisense DNA probes against Linc00632 for biotin pull-down assay, followed by qRT-PCR analysis to examine miR-498 levels. g Lysates from NECs were incubated with in vitro-synthesized biotin-labeled Linc00632 and antisense RNA for biotin pull-down assay, followed by real-time-PCR analysis to examine miR-498 levels. h Correlation analysis between Linc00632 and miR-498 in mucosal tissues from 30 AR patients. Data are representative of 3 or more independent experiments. * p < 0.05; ** p < 0.01; *** p < 0.001. NECs, nasal epithelial cells; ns, not significant.