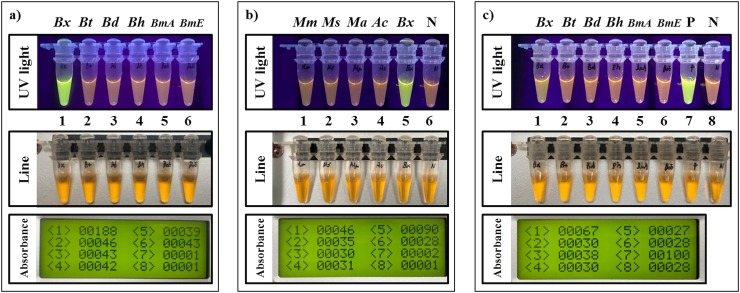
Fig 4. Three types of specificity tests of the RPA assay for B. xylophilus.
a) The specificity test of the RPA assay using 6 ng of pure gDNA from B. xylophilus (Bx) and five Bursaphelenchus spp.: B. thailandae (Bt), B. doui (Bd), B. hylobianum (Bh), B. mucronatus Asia type (BmA), and B. mucronatus Europe type (BmE) which have strong genetic relationships with B. xylophilus. b) The specificity test of the RPA assay on organisms: Matsucoccus matsumurae (Mm), Monochamus saltuarius (Ms), Monochamus alternatus (Ma), and Anoplophora chinensis (Ac) which have weak genetic relationships with B. xylophilus. c) The specificity test of the RPA assay using spiked pinewood from healthy pine tree with pure gDNA from nematodes. The absorbance and UV light represent the detection results of the POID and the UV light, respectively. P and N represent the positive control using pure gDNA from B. xylophilus and the negative control using lysate from healthy pine tree extracted using the DAP buffer, respectively. The line numbering is matched with the absorbance numbering in each panel. The absorbance value of the zero point in the RPA assay was set to 50. Therefore, values of less than 50 measured by the POID are considered not specifically amplified.