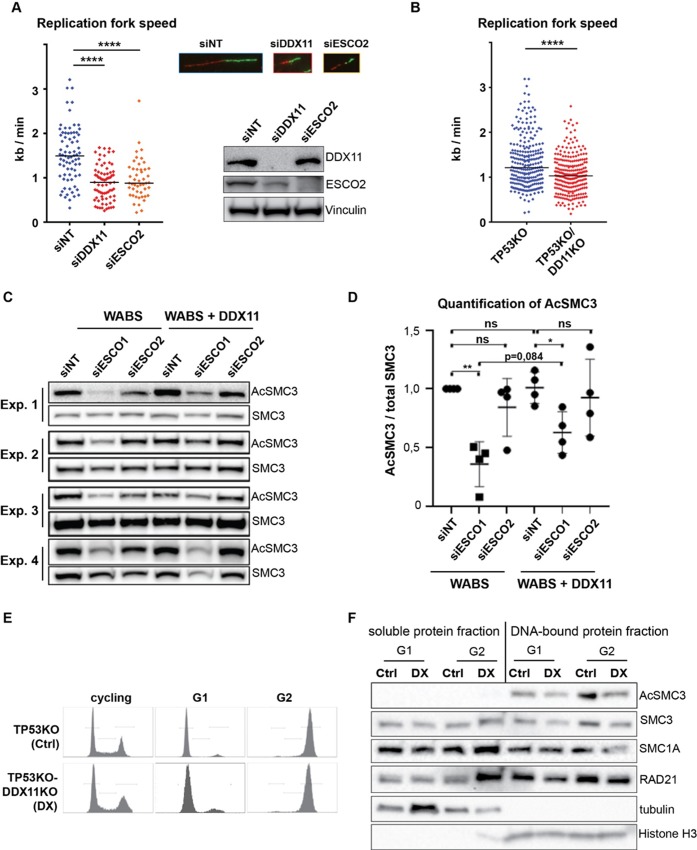
Fig 7. DDX11 deficiency slows replication fork speed and reduces SMC3 acetylation.
(A) RPE1-hTERT cells were transfected with indicated siRNAs for two days and analyzed by western blot. Replication fork speed was assessed with a DNA fiber assay using a double labeling protocol. Black lines indicate the median. P-values were calculated by a non-parametric one-way ANOVA test. **** p<0,0001. Representative images of DNA fiber tracts are shown. (B) RPE1-TP53KO and RPE1-TP53KO-DDX1KO cells were analyzed with a DNA fiber assay. (C) WABS fibroblasts and corrected cells were transfected with the indicated siRNAs and analyzed by western blot after three days. Additional antibody incubations are provided in S2 Fig. (D) Quantification of AcSMC3 levels normalized to total SMC3, using image-lab software and. Mean and standard deviations are shown. P-values were calculated using a two-tailed t-test. * p<0,05; **<0,01; ns not significant. (E) RPE1-TP53KO and RPE1-TP53KO-DDX11KO cells were arrested in G1 using the Cdk4/6 inhibitor Palbociclib (20h, 10 μM) and in G2 using the Cdk1 inhibitor RO3306 (20h, 10 μM) and assessed by flow cytometry. (F) cells from (E) were lysed, and soluble and DNA-bound protein fractions were separated and analyzed by western blot. Ctrl, TP53KO cells; DX, TP53KO-DDX11KO cells.