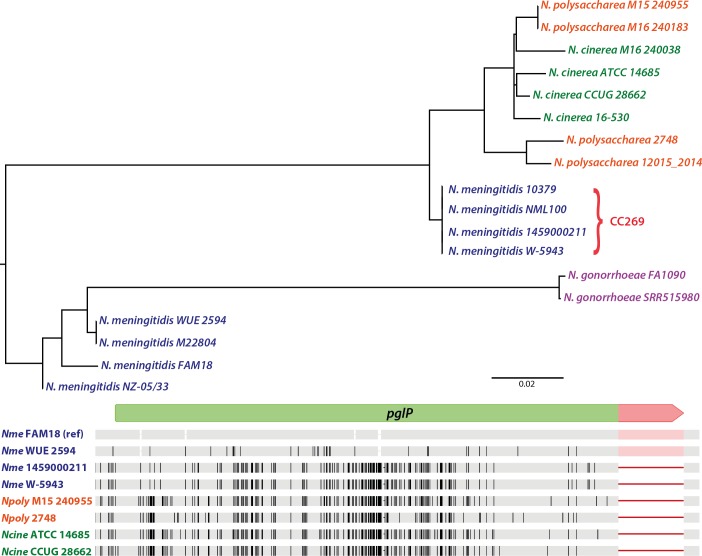
Fig 7. HGT-mediated acquisition of an intact pglP allele in CC269 strains.
ORF nucleotide sequences from pglP alleles (excluding the ORF—extending CREE insertion) were aligned for phylogenetic sequence analyses using Clustal W (top panel). Multiple-alignment view of the variant nucleotide sites detected in pglP with that of FAM18 set as a reference sequence (bottom panel). The FAM18 allele contains the C-terminal, ORF—extending CREE insertion (segment in pink). Each SNP that differed from FAM18 is shown as a single line with thicker lines indicative of multiple neighboring SNPs. The sequence identities of another N. meningitidis pglP pseudogene, two CC269 alleles, two N. polysaccharea alleles and two N. cinerea alleles generated using ParSNP are shown. Note the absence of the CREE sequences in the CC269, N. polysaccharea and N. cinerea alleles and that greater than 80% of all pglP alleles in CC269 strains are 100% identical to those identified here.