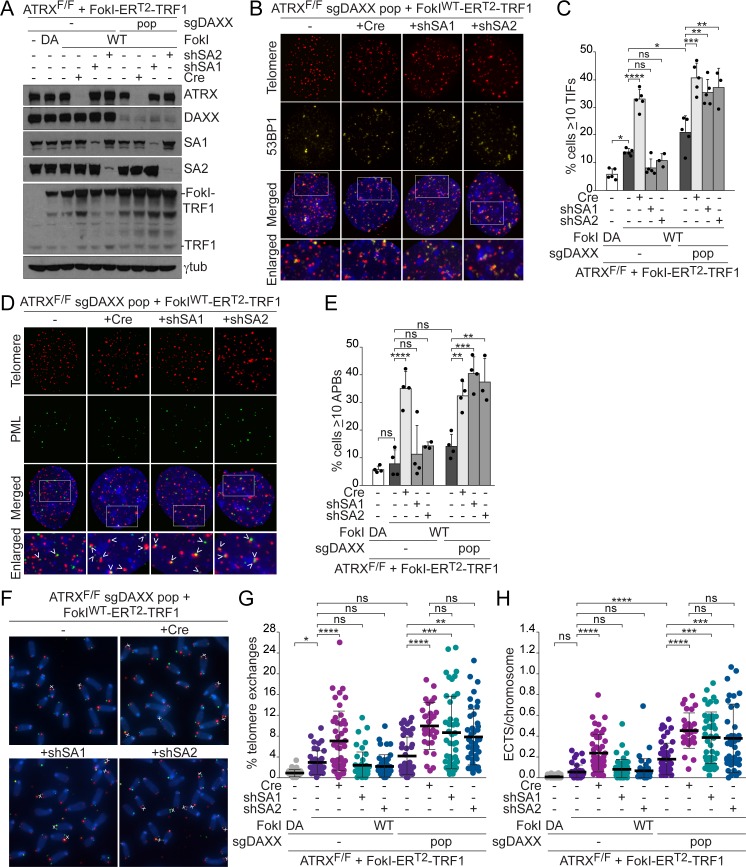
Fig 6. The combined loss of DAXX and telomere cohesion phenocopies ATRX loss.
(A) Immunoblots showing FokI-ERT2-TRF1 expression (6 days) in control and a CRISPR/Cas9-targeted DAXX-deficient population (pop) of ATRXF/F MEFs. Cre-mediated deletion of ATRX and shRNA knockdown of SA1 and SA2 are also shown (4 days). γtubulin serves as a loading control. (B) Detection of TIFs in DAXX-targeted cells expressing FokIWT-ERT2-TRF1 by IF-FISH for 53BP1 and telomeres as in Fig 1C. (C) Quantification of the TIF response in FokI-ERT2-TRF1–expressing cells, as assayed in (B). Bars: means and SDs from at least 3 experiments of >100 cells each. (D) APBs in DAXX-targeted MEFs expressing FokIWT-ERT2-TRF1 detected by IF-FISH as in Fig 1E. (E) Quantification of the percentage of cells with ≥10 APBs in cells expressing FokI-ERT2-TRF1, as assayed in (D). Bars: means and SDs from at least 3 experiments of >100 cells each. (F) CO-FISH staining of metaphase spreads from DAXX-targeted ATRXF/F MEFs expressing FokIWT-ERT2-TRF1 as in Fig 1G. (G) Quantification of telomere exchanges detected by CO-FISH in control and DAXX-deficient ATRXF/F MEFs expressing FokI-ERT2-TRF1. Each data point represents the percentage of chromosome ends with telomere exchanges in one metaphase spread. Bars: means and SDs of >40 metaphases from 5 experiments (29 metaphases from 3 experiments for sgDAXX pop+Cre). (H) Quantification of the ECTSs from metaphase spreads described in (F, G). Each data point represents the number of ECTSs per chromosome in one metaphase spread. All p-values were derived from a one-way ANOVA with Tukey correction. Symbols as in Fig 1. The underlying numerical data and statistical analysis for each figure panel can be found in S1 Data. APB, ALT-associated PML body; ATRX, alpha thalassemia/mental retardation syndrome X-linked chromatin remodeler; ATRXF/F, female embryo with two floxed ATRX alleles; Cas9, CRISPR associated protein 9; CO-FISH, chromosome orientation fluorescence in situ hybridization; Cre, recombinase acting on Lox sites; CRISPR, clustered regularly interspaced short palindromic repeats; DA, nuclease dead FokI-ERT2-TRF1; DAXX, death domain-associated protein; ECTS, extrachromosomal telomeric signal; FISH, fluorescence in situ hybridization; FokI-ERT2-TRF1, tamoxifen-inducible nuclear localized FokI-TRF1 fusion protein; IF, immunofluorescence; MEF, mouse embryonic fibroblast; ns, not significant; PML, promyelocytic leukemia; SA1, stromal antigen 1; SA2, stromal antigen 2; SD, standard deviation; sgDAXX, single guide RNA for DAXX; shRNA, short hairpin RNA; shSA1, short hairpin RNA for SA1; shSA2, short hairpin RNA for SA2; TIF, telomere dysfunction–induced foci; WT, wild-type FokI-ERT2-TRF1; 53BP1, tumor protein p53-binding protein 1.