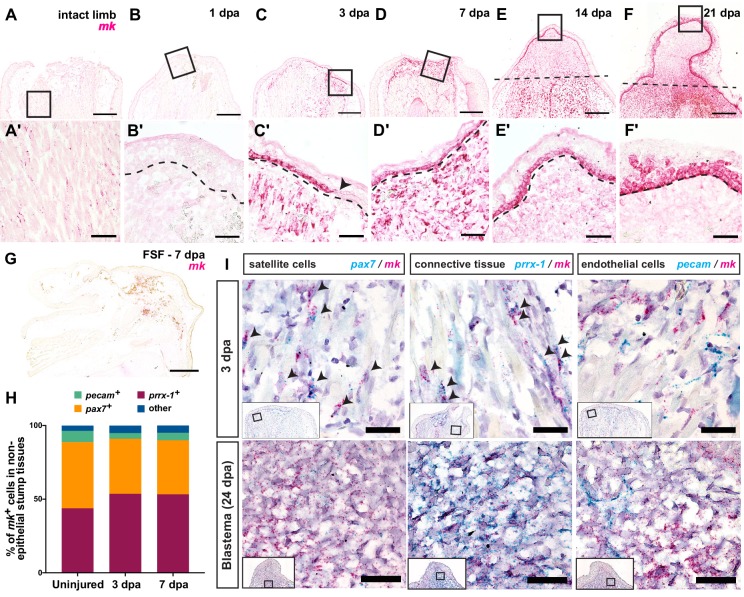
Figure 2. Midkine (mk) is highly expressed in the basal layers of the wound epidermis/AEC and blastemal progenitors.
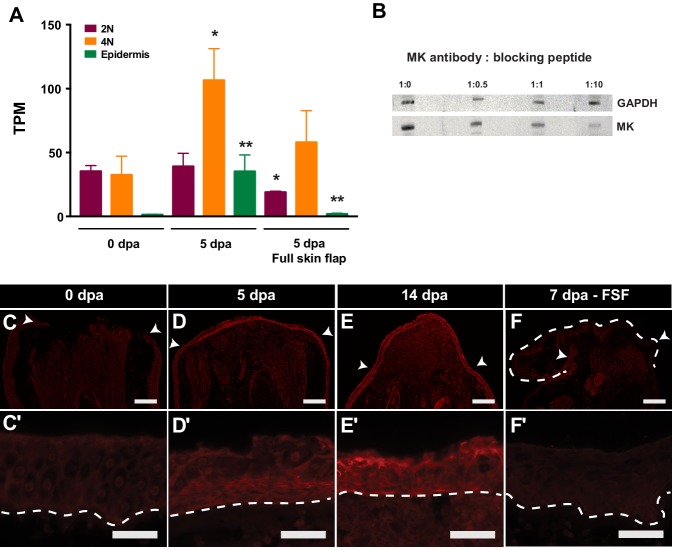
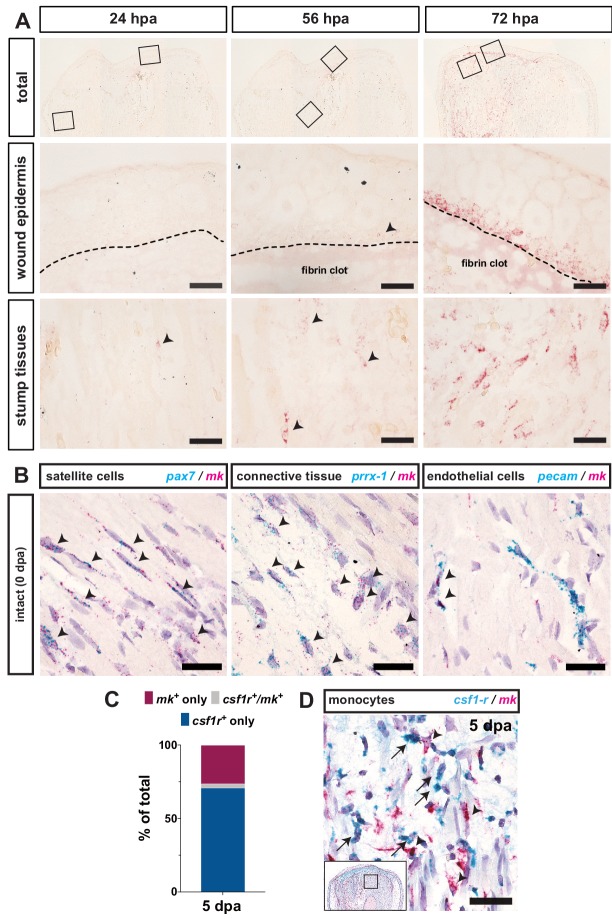
(A–F’) Timecourse RNAscope in situ hybridization of midkine at 0 (intact), 1, 3, 7, 14, and 21 dpa. Insets in A-F are shown in A’-F’ at higher magnification. Arrowhead in C’ denotes the beginning of the wound epidermis. Dotted line marks amputation plane in E and F or wound epidermis/AEC boundary in B’-F’. (G) In situ hybridization of mk in full skin flap sutured limbs at 7 dpa. Axolotl MK protein expression can be found in Figure 2—figure supplement 1. (H) Breakdown of the percentages of mk+ cells that are pax7+, prrx-1+, and pecam+ in regenerating stump tissues during early stages of regeneration. At 3 dpa, a total N of 1579, 444, and 1180 cells were counted for pax7, prrx-1, and pecam double in situs, respectively. At 7 dpa, a total N of 456, 1043, and 274 cells were counted for pax7, prrx-1, and pecam double in situs, respectively (Figure 2—source data 1). (I) Representative images of RNAscope double in situ hybridization of mk with pax7 (left), prrx-1 (middle), or pecam (right) at early (3 dpa) and later blastema (24 dpa) stages. Insets depict where higher magnification images were taken. Black arrowheads mark double positive cells. More detailed analysis of the onset of mk expression in early stages of regeneration, representative images of double in situ hybridization of mk with cell type-specific markers in uninjured tissue, as well as the analysis of mk co-expression with the monocyte marker csf1r can be found in Figure 2—figure supplement 2. Scale bars, A-G: 500 µm, A’-F’: 100 µm, I: 50 µm. dpa, days post-amputation, FSF, full skin flap.