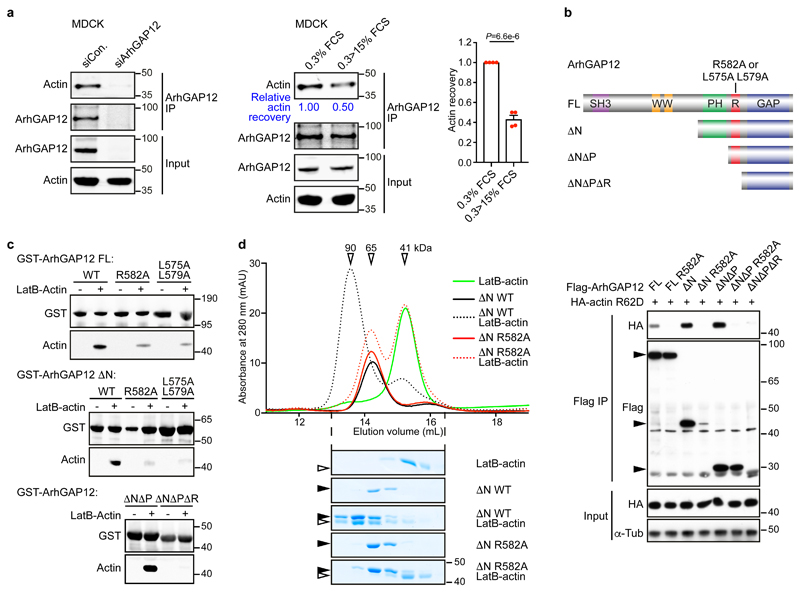
Figure 2. ArhGAP12 interaction with G-actin requires the RPEL motif.
(a) Endogenous ArhGAP12 was immunoprecipitated and actin recovery was analysed by immunoblot in starved MDCK cells. Cells were transfected with control or ArhGAP12 siRNA (left; data shown represent 4 independent experiments), or serum-stimulated as indicated (middle and right). Bar graph data are mean ± SEM, n=4 independent experiments, two-tailed unpaired t-test. (b) Top, ArhGAP12 derivatives: full-length (FL), amino acids 1-791; ΔN 410-791; ΔNΔP 568-791; ΔNΔPΔR, 582-791. RPEL point mutants were R582A and L575A L579A. Bottom, nonpolymerisable actin mutant R62D was coexpressed with wildtype ArhGAP12 or RPEL mutant R582A in NIH3T3 cells and their interaction analysed by immunoprecipitation and immunoblotting. (c) Immobilised recombinant GST-ArhGAP12 proteins were used to recover purified LatB-actin from solution; actin recovery was analysed by immunoblot. (d) Analytical gel filtration. Elution profiles of recombinant ArhGAP12 ΔN (4 μM) and purified LatB-actin (5 μM) either alone (solid lines) or in a mixture (dotted lines), analysed by absorbance (top) or SDS-PAGE/Coomassie blue staining (bottom). Apparent Mr are indicated. Black and open horizontal arrowheads point to ArhGAP12 and actin respectively. Data shown in (b-d) are representative of 3 independent experiments, respectively. See Supplementary Figure 1 for related data. Source data for a and d are shown in Supplementary Table 1. Unprocessed blots and Coomassie gels are shown in Supplementary Figure 8.